The transcriptomes of the cattle parasitic nematode Ostertagia ostartagi
- PMID: 19346077
- PMCID: PMC2677129
- DOI: 10.1016/j.vetpar.2009.02.023
The transcriptomes of the cattle parasitic nematode Ostertagia ostartagi
Abstract
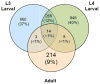
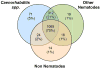
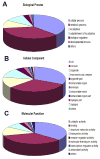
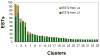
Ostertagia ostertagi is a gastrointestinal parasitic nematode that affects cattle and leads to a loss of production. In this study, we present the first large-scale genomic survey of O. ostertagi by the analysis of expressed transcripts from three stages of the parasite: third-stage larvae, fourth-stage larvae and adult worms. Using an in silico approach, 2284 genes were identified from over 7000 expressed sequence tags and abundant transcripts were analyzed and characterized by their functional profile. Of the 2284 genes, 66% had similarity to other known or predicted genes while the rest were novel and potentially represent genes specific to the species and/or stages. Furthermore, a subset of the novel proteins were structurally annotated and assigned putative function based on orthologs in Caenorhabditis elegans and corresponding RNA interference phenotypes. Hence, over 70% of the genes were annotated using protein sequences, domains and pathway databases. Differentially expressed transcripts from the two larval stages and their functional profiles were also studied leading to a more detailed understanding of the parasite's life-cycle. The identified transcripts are a valuable resource for genomic studies of O. ostertagi and can facilitate the design of control strategies and vaccine programs.
Figures

Similar articles
-
Transcriptome analyses reveal protein and domain families that delineate stage-related development in the economically important parasitic nematodes, Ostertagia ostertagi and Cooperia oncophora.BMC Genomics. 2013 Feb 22;14:118. doi: 10.1186/1471-2164-14-118. BMC Genomics. 2013. PMID: 23432754 Free PMC article.
-
Analysis of the transthyretin-like (TTL) gene family in Ostertagia ostertagi--comparison with other strongylid nematodes and Caenorhabditis elegans.Int J Parasitol. 2008 Nov;38(13):1545-56. doi: 10.1016/j.ijpara.2008.04.004. Epub 2008 May 10. Int J Parasitol. 2008. PMID: 18571174
-
Efficacy and specificity of RNA interference in larval life-stages of Ostertagia ostertagi.Parasitology. 2006 Dec;133(Pt 6):777-83. doi: 10.1017/S0031182006001004. Epub 2006 Jul 31. Parasitology. 2006. PMID: 16879764
-
Altered avr-14B gene transcription patterns in ivermectin-resistant isolates of the cattle parasites, Cooperia oncophora and Ostertagia ostertagi.Int J Parasitol. 2011 Aug 1;41(9):951-7. doi: 10.1016/j.ijpara.2011.04.003. Epub 2011 Jun 1. Int J Parasitol. 2011. PMID: 21683704
-
Role of the bovine immune system and genome in resistance to gastrointestinal nematodes.Vet Parasitol. 2001 Jul 12;98(1-3):51-64. doi: 10.1016/s0304-4017(01)00423-x. Vet Parasitol. 2001. PMID: 11516579 Review.
Cited by
-
Assembly and Analysis of Haemonchus contortus Transcriptome as a Tool for the Knowledge of Ivermectin Resistance Mechanisms.Pathogens. 2023 Mar 22;12(3):499. doi: 10.3390/pathogens12030499. Pathogens. 2023. PMID: 36986421 Free PMC article.
-
Transcriptome analyses reveal protein and domain families that delineate stage-related development in the economically important parasitic nematodes, Ostertagia ostertagi and Cooperia oncophora.BMC Genomics. 2013 Feb 22;14:118. doi: 10.1186/1471-2164-14-118. BMC Genomics. 2013. PMID: 23432754 Free PMC article.
-
Proteomic analysis of secretory products from the model gastrointestinal nematode Heligmosomoides polygyrus reveals dominance of venom allergen-like (VAL) proteins.J Proteomics. 2011 Aug 24;74(9):1573-94. doi: 10.1016/j.jprot.2011.06.002. Epub 2011 Jun 29. J Proteomics. 2011. PMID: 21722761 Free PMC article.
-
An analysis of the transcriptome of Teladorsagia circumcincta: its biological and biotechnological implications.BMC Genomics. 2012;13 Suppl 7(Suppl 7):S10. doi: 10.1186/1471-2164-13-S7-S10. Epub 2012 Dec 13. BMC Genomics. 2012. PMID: 23282110 Free PMC article.
-
The identification of small molecule inhibitors with anthelmintic activities that target conserved proteins among ruminant gastrointestinal nematodes.mBio. 2024 Mar 13;15(3):e0009524. doi: 10.1128/mbio.00095-24. Epub 2024 Feb 15. mBio. 2024. PMID: 38358246 Free PMC article.
References
-
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. - PMC - PubMed
-
- Audic S, Claverie JM. The Significance of Digital Gene Expression Profiles. Genome Res. 1997;7:986–995. - PubMed
-
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, Vida JT, Thomas WK. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. - PubMed
-
- Burnell AM, Houthoofd K, O’Hanlon K, Vanfleteren JR. Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans. Exp Gerontol. 2005;40:850–856. - PubMed
-
- Elling A, Mitreva M, Recknor J, Gai X, Martin J, Maier T, McDermott J, Hewezi T, McK Bird D, Davis E, Hussey R, Nettleton D, McCarter J, Baum T. Divergent evolution of arrested development in the dauer stage of Caenorhabditis elegans and the infective stage of Heterodera glycines. Genome Biol. 2007;8:R211. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

