Characterization of the DraT/DraG system for posttranslational regulation of nitrogenase in the endophytic betaproteobacterium Azoarcus sp. strain BH72
- PMID: 19346301
- PMCID: PMC2681912
- DOI: 10.1128/JB.01720-08
Characterization of the DraT/DraG system for posttranslational regulation of nitrogenase in the endophytic betaproteobacterium Azoarcus sp. strain BH72
Abstract
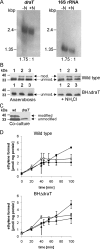
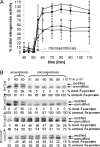
DraT/DraG-mediated posttranslational regulation of the nitrogenase Fe protein by ADP-ribosylation has been described for a few diazotrophic bacteria belonging to the class Alphaproteobacteria. Here we present for the first time the DraT/DraG system of a betaproteobacterium, Azoarcus sp. strain BH72, a diazotrophic grass endophyte. Its genome harbors one draT ortholog and two physically unlinked genes coding for ADP-ribosylhydrolases. Northern blot analysis revealed cotranscription of draT with two genes encoding hypothetical proteins. Furthermore, draT and draG2 were expressed under all studied conditions, whereas draG1 expression was nitrogen regulated. By using Western blot analysis of deletion mutants and nitrogenase assays in vivo, we demonstrated that DraT is required for the nitrogenase Fe protein modification but not for the physiological inactivation of nitrogenase activity. A second mechanism responsible for nitrogenase inactivation must operate in this bacterium, which is independent of DraT. Fe protein demodification was dependent mainly on DraG1, corroborating the assumption from phylogenetic analysis that DraG2 might be mostly involved in processes other than the posttranslational regulation of nitrogenase. Nitrogenase in vivo reactivation was impaired in a draG1 mutant and a mutant lacking both draG alleles after anaerobiosis shifts and subsequent adjustment to microaerobic conditions, suggesting that modified dinitrogenase reductase was inactive. Our results demonstrate that the DraT/DraG system, despite some differences, is functionally conserved in diazotrophic proteobacteria.
Figures


References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
-
- Dean, D. R., and M. R. Jacobson. 1992. Biochemical genetics of nitrogenase, p. 763-835. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, NY.
-
- Durner, J., I. Böhm, H. Hilz, and P. Böger. 1994. Posttranslational modification of nitrogenase. Differences between the purple bacterium Rhodospirillum rubrum and the cyanobacterium Anabaena variabilis. Eur. J. Biochem. 220125-130. - PubMed
-
- Edgren, T., and S. Nordlund. 2005. Electron transport to nitrogenase in Rhodospirillum rubrum: identification of a new fdxN gene encoding the primary electron donor to nitrogenase. FEMS Microbiol. Lett. 245345-351. - PubMed
-
- Egener, T., T. Hurek, and B. Reinhold-Hurek. 1999. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant-Microbe Interact. 12813-819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

