Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors
- PMID: 19346308
- PMCID: PMC2681890
- DOI: 10.1128/JB.01824-08
Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors
Abstract
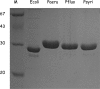
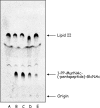
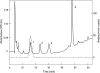
Genes encoding proteins that exhibit similarity to the C-terminal domain of Escherichia coli colicin M were identified in the genomes of some Pseudomonas species, namely, P. aeruginosa, P. syringae, and P. fluorescens. These genes were detected only in a restricted number of strains. In P. aeruginosa, for instance, the colicin M homologue gene was located within the ExoU-containing genomic island A, a large horizontally acquired genetic element and virulence determinant. Here we report the cloning of these genes from the three Pseudomonas species and the purification and biochemical characterization of the different colicin M homologues. All of them were shown to exhibit Mg(2+)-dependent diphosphoric diester hydrolase activity toward the two undecaprenyl phosphate-linked peptidoglycan precursors (lipids I and II) in vitro. In all cases, the site of cleavage was localized between the undecaprenyl and pyrophospho-MurNAc moieties of these precursors. These enzymes were not active on the cytoplasmic precursor UDP-MurNAc-pentapeptide or (or only very poorly) on undecaprenyl pyrophosphate. These colicin M homologues have a narrow range of antibacterial activity. The P. aeruginosa protein at low concentrations was shown to inhibit growth of sensitive P. aeruginosa strains. These proteins thus represent a new class of bacteriocins (pyocins), the first ones reported thus far in the genus Pseudomonas that target peptidoglycan metabolism.
Figures


References
-
- Amrein, K. E., B. Takacs, M. Stieger, J. Molnos, N. A. Flint, and P. Burn. 1995. Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc. Natl. Acad. Sci. USA 921048-1052. - PMC - PubMed
-
- Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 4473-88. - PubMed
-
- Bouhss, A., M. Crouvoisier, D. Blanot, and D. Mengin-Lecreulx. 2004. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 27929974-29980. - PubMed
-
- Bouhss, A., A. E. Trunkfield, T. D. Bugg, and D. Mengin-Lecreulx. 2008. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32208-233. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

