Structure-based design of a periplasmic binding protein antagonist that prevents domain closure
- PMID: 19348466
- PMCID: PMC2742562
- DOI: 10.1021/cb900021q
Structure-based design of a periplasmic binding protein antagonist that prevents domain closure
Abstract
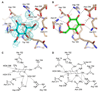
Many receptors undergo ligand-induced conformational changes to initiate signal transduction. Periplasmic binding proteins (PBPs) are bacterial receptors that exhibit dramatic conformational changes upon ligand binding. These proteins mediate a wide variety of fundamental processes including transport, chemotaxis, and quorum sensing. Despite the importance of these receptors, no PBP antagonists have been identified and characterized. In this study, we identify 3-O-methyl-d-glucose as an antagonist of glucose/galactose-binding protein and demonstrate that it inhibits glucose chemotaxis in E. coli. Using small-angle X-ray scattering and X-ray crystallography, we show that this antagonist acts as a wedge. It prevents the large-scale domain closure that gives rise to the active signaling state. Guided by these results and the structures of open and closed glucose/galactose-binding protein, we designed and synthesized an antagonist composed of two linked glucose residues. These findings provide a blueprint for the design of new bacterial PBP inhibitors. Given the key role of PBPs in microbial physiology, we anticipate that PBP antagonists will have widespread uses as probes and antimicrobial agents.
Figures




References
-
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. - PubMed
-
- Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 1996;20:17–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

