The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization
- PMID: 19348770
- PMCID: PMC3325131
- DOI: 10.1016/j.bpj.2008.12.3942
The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization
Abstract
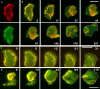
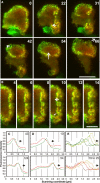
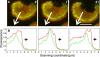
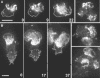
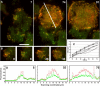
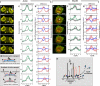
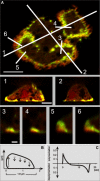
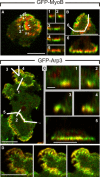
Actin polymerization is typically initiated at specific sites in a cell by membrane-bound protein complexes, and the resulting structures are involved in specialized cellular functions, such as migration, particle uptake, or mitotic division. Here we analyze the potential of the actin system to self-organize into waves that propagate on the planar, substrate-attached membrane of a cell. We show that self-assembly involves the ordered recruitment of proteins from the cytoplasmic pool and relate the organization of actin waves to their capacity for applying force. Three proteins are shown to form distinct three-dimensional patterns in the actin waves. Myosin-IB is enriched at the wave front and close to the plasma membrane, the Arp2/3 complex is distributed throughout the waves, and coronin forms a sloping layer on top of them. CARMIL, a protein that links myosin-IB to the Arp2/3 complex, is also recruited to the waves. Wave formation does not depend on signals transmitted by heterotrimeric G-proteins, nor does their propagation require SCAR, a regulator upstream of the Arp2/3 complex. Propagation of the waves is based on an actin treadmilling mechanism, indicating a program that couples actin assembly to disassembly in a three-dimensional pattern. When waves impinge on the cell perimeter, they push the edge forward; when they reverse direction, the cell border is paralyzed. These data show that force-generating, highly organized supramolecular networks are autonomously formed in live cells from molecular motors and proteins controlling actin polymerization and depolymerization.
Figures


References
-
- Vicker M.G. Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp. Cell Res. 2002;275:54–66. - PubMed
-
- Bretschneider T., Diez S., Anderson K., Heuser J., Clarke M. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 2004;14:1–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

