A novel mechanism of antagonism between ATP-dependent chromatin remodeling complexes regulates RNR3 expression
- PMID: 19349301
- PMCID: PMC2698743
- DOI: 10.1128/MCB.01741-08
A novel mechanism of antagonism between ATP-dependent chromatin remodeling complexes regulates RNR3 expression
Abstract
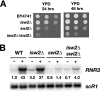
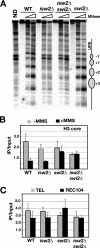
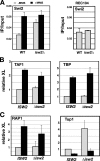
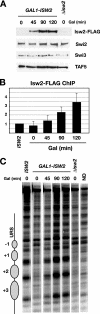
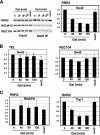
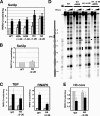
Gene expression depends upon the antagonistic actions of chromatin remodeling complexes. While this has been studied extensively for the enzymes that covalently modify the tails of histones, the mechanism of how ATP-dependent remodeling complexes antagonize each other to maintain the proper level of gene activity is not known. The gene encoding a large subunit of ribonucleotide reductase, RNR3, is regulated by ISW2 and SWI/SNF, complexes that repress and activate transcription, respectively. Here, we studied the functional interactions of these two complexes at RNR3. Deletion of ISW2 causes constitutive recruitment of SWI/SNF, and conditional reexpression of ISW2 causes the repositioning of nucleosomes and reduced SWI/SNF occupancy at RNR3. Thus, ISW2 is required for restriction of access of SWI/SNF to the RNR3 promoter under the uninduced condition. Interestingly, the binding of sequence-specific DNA binding factors and the general transcription machinery are unaffected by the status of ISW2, suggesting that disruption of nucleosome positioning does not cause a nonspecific increase in cross-linking of all factors to RNR3. We provide evidence that ISW2 does not act on SWI/SNF directly but excludes its occupancy by positioning nucleosomes over the promoter. Genetic disruption of nucleosome positioning by other means led to a similar phenotype, linking repressed chromatin structure to SWI/SNF exclusion. Thus, incorporation of promoters into a repressive chromatin structure is essential for prevention of the opportunistic actions of nucleosome-disrupting activities in vivo, providing a novel mechanism for maintaining tight control of gene expression.
Figures

References
-
- Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111381-392. - PubMed
-
- Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. - PubMed
-
- Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10935-942. - PubMed
-
- Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4715-723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
