Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability
- PMID: 19349461
- PMCID: PMC2715118
- DOI: 10.1084/jem.20082396
Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability
Abstract
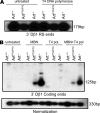
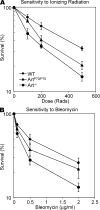
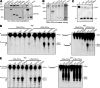
Artemis was initially discovered as the gene inactivated in human radiosensitive T(-)B(-) severe combined immunodeficiency, a syndrome characterized by the absence of B and T lymphocytes and cellular hypersensitivity to ionizing radiation. Hypomorphic Artemis alleles have also been identified in patients and are associated with combined immunodeficiencies of varying severity. We examine the molecular mechanisms underlying a syndrome of partial immunodeficiency caused by a hypomorphic Artemis allele using the mouse as a model system. This mutation, P70, leads to premature translation termination that deletes a large portion of a nonconserved C terminus. We find that homozygous Artemis-P70 mice exhibit reduced numbers of B and T lymphocytes, thereby recapitulating the patient phenotypes. The hypomorphic mutation results in impaired end processing during the lymphoid-specific DNA rearrangement known as V(D)J recombination, defective double-strand break repair, and increased chromosomal instability. Biochemical analyses reveal that the Artemis-P70 mutant protein interacts with the DNA-dependent protein kinase catalytic subunit and retains significant, albeit reduced, exo- and endonuclease activities but does not undergo phosphorylation. Together, our findings indicate that the Artemis C terminus has critical in vivo functions in ensuring efficient V(D)J rearrangements and maintaining genome integrity.
Figures



References
-
- Bassing C.H., Swat W., Alt F.W. 2002. The mechanism and regulation of chromosomal V(D)J recombination.Cell. 109:S45–S55 - PubMed
-
- Sekiguchi J., Alt F.W., Oettinger M. 2004. The mechanism of V(D)J recombination. Molecular Biology of B cells. Alt F.W., Honjo T., Academic Press, San Diego, CA: 57–78
-
- Fugmann S.D. 2001. RAG1 and RAG2 in V(D)J recombination and transposition.Immunol. Res. 23:23–39 - PubMed
-
- Gellert M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation.Annu. Rev. Biochem. 71:101–132 - PubMed
-
- Buck D., Malivert L., de Chasseval R., Barraud A., Fondaneche M.C., Sanal O., Plebani A., Stephan J.L., Hufnagel M., le Deist F., et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly.Cell. 124:287–299 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

