Control of T helper cell differentiation through cytokine receptor inclusion in the immunological synapse
- PMID: 19349465
- PMCID: PMC2715121
- DOI: 10.1084/jem.20082900
Control of T helper cell differentiation through cytokine receptor inclusion in the immunological synapse
Abstract
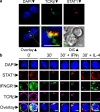
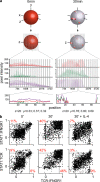
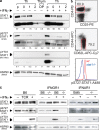
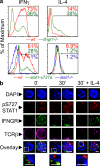
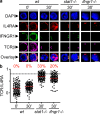
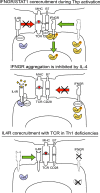
The antigen recognition interface formed by T helper precursors (Thps) and antigen-presenting cells (APCs), called the immunological synapse (IS), includes receptors and signaling molecules necessary for Thp activation and differentiation. We have recently shown that recruitment of the interferon-gamma receptor (IFNGR) into the IS correlates with the capacity of Thps to differentiate into Th1 effector cells, an event regulated by signaling through the functionally opposing receptor to interleukin-4 (IL4R). Here, we show that, similar to IFN-gamma ligation, TCR stimuli induce the translocation of signal transducer and activator of transcription 1 (STAT1) to IFNGR1-rich regions of the membrane. Unexpectedly, STAT1 is preferentially expressed, is constitutively serine (727) phosphorylated in Thp, and is recruited to the IS and the nucleus upon TCR signaling. IL4R engagement controls this process by interfering with both STAT1 recruitment and nuclear translocation. We also show that in cells with deficient Th1 or constitutive Th2 differentiation, the IL4R is recruited to the IS. This observation suggest that the IL4R is retained outside the IS, similar to the exclusion of IFNGR from the IS during IL4R signaling. This study provides new mechanistic cues for the regulation of lineage commitment by mutual immobilization of functionally antagonistic membrane receptors.
Figures


References
-
- Szabo S.J., Sullivan B.M., Peng S.L., Glimcher L.H. 2003. Molecular mechanisms regulating Th1 immune responses.Annu. Rev. Immunol. 21:713–758 - PubMed
-
- Murphy K.M., Reiner S.L. 2002. The lineage decisions of helper T cells.Nat. Rev. Immunol. 2:933–944 - PubMed
-
- Davis D.M., Dustin M.L. 2004. What is the importance of the immunological synapse? Trends Immunol. 25:323–327 - PubMed
-
- Friedl P., den Boer A.T., Gunzer M. 2005. Tuning immune responses: diversity and adaptation of the immunological synapse.Nat. Rev. Immunol. 5:532–545 - PubMed
-
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., Dustin M.L. 1999. The immunological synapse: a molecular machine controlling T cell activation.Science. 285:221–227 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

