Molecular signatures of cell migration in C. elegans Q neuroblasts
- PMID: 19349580
- PMCID: PMC2700521
- DOI: 10.1083/jcb.200812077
Molecular signatures of cell migration in C. elegans Q neuroblasts
Abstract
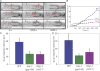
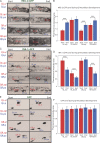
Metazoan cell movement has been studied extensively in vitro, but cell migration in living animals is much less well understood. In this report, we have studied the Caenorhabditis elegans Q neuroblast lineage during larval development, developing live animal imaging methods for following neuroblast migration with single cell resolution. We find that each of the Q descendants migrates at different speeds and for distinct distances. By quantitative green fluorescent protein imaging, we find that Q descendants that migrate faster and longer than their sisters up-regulate protein levels of MIG-2, a Rho family guanosine triphosphatase, and/or down-regulate INA-1, an integrin alpha subunit, during migration. We also show that Q neuroblasts bearing mutations in either MIG-2 or INA-1 migrate at reduced speeds. The migration defect of the mig-2 mutants, but not ina-1, appears to result from a lack of persistent polarization in the direction of cell migration. Thus, MIG-2 and INA-1 function distinctly to control Q neuroblast migration in living C. elegans.
Figures



References
-
- Baum P.D., Garriga G. 1997. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants.Neuron. 19:51–62 - PubMed
-
- Branda C.S., Stern M.J. 2000. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites.Dev. Biol. 226:137–151 - PubMed
-
- Cordes S., Frank C.A., Garriga G. 2006. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions.Development. 133:2747–2756 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

