Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells
- PMID: 19350678
- PMCID: PMC4357362
- DOI: 10.1002/stem.13
Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells
Abstract
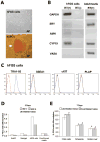
The derivation of germ cells from human embryonic stem cells (hESCs) or human induced pluripotent stem (hIPS) cells represents a desirable experimental model and potential strategy for treating infertility. In the current study, we developed a triple biomarker assay for identifying and isolating human primordial germ cells (PGCs) by first evaluating human PGC formation during the first trimester in vivo. Next, we applied this technology to characterizing in vitro derived PGCs (iPGCs) from pluripotent cells. Our results show that codifferentiation of hESCs on human fetal gonadal stromal cells significantly improves the efficiency of generating iPGCs. Furthermore, the efficiency was comparable between various pluripotent cell lines regardless of origin from the inner cell mass of human blastocysts (hESCs), or reprogramming of human skin fibroblasts (hIPS). To better characterize the iPGCs, we performed Real-time polymerase chain reaction, microarray, and bisulfite sequencing. Our results show that iPGCs at day 7 of differentiation are transcriptionally distinct from the somatic cells, expressing genes associated with pluripotency and germ cell development while repressing genes associated with somatic differentiation (specifically multiple HOX genes). Using bisulfite sequencing, we show that iPGCs initiate imprint erasure from differentially methylated imprinted regions by day 7 of differentiation. However, iPGCs derived from hIPS cells do not initiate imprint erasure as efficiently. In conclusion, our results indicate that triple positive iPGCs derived from pluripotent cells differentiated on hFGS cells correspond to committed first trimester germ cells (before 9 weeks) that have initiated the process of imprint erasure.
Figures





References
-
- Daley GQ. Gametes from embryonic stem cells: a cup half empty or half full? Science. 2007 Apr 20;316(5823):409–410. - PubMed
-
- Holden C. Sperm from Skin Becoming a Reality. ScienceNOW daily News. 2008;415:2.
-
- Surani MA. Germ cells: the eternal link between generations. C R Biol. 2007 Jun-Jul;330(6-7):474–478. - PubMed
-
- Hayashi K, De Sousa Lopes S, Surani M. Germ cell specification in mice. Science. 2007;316:394–396. - PubMed
-
- Ying Y, L XM, Marble A, Lawson KA, Zhao GQ. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol. 2000;14:1053–1063. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

