IMGD: an integrated platform supporting comparative genomics and phylogenetics of insect mitochondrial genomes
- PMID: 19351385
- PMCID: PMC2681477
- DOI: 10.1186/1471-2164-10-148
IMGD: an integrated platform supporting comparative genomics and phylogenetics of insect mitochondrial genomes
Abstract
Background: Sequences and organization of the mitochondrial genome have been used as markers to investigate evolutionary history and relationships in many taxonomic groups. The rapidly increasing mitochondrial genome sequences from diverse insects provide ample opportunities to explore various global evolutionary questions in the superclass Hexapoda. To adequately support such questions, it is imperative to establish an informatics platform that facilitates the retrieval and utilization of available mitochondrial genome sequence data.
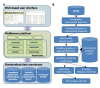
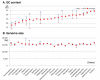
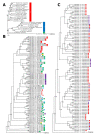
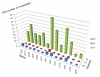
Results: The Insect Mitochondrial Genome Database (IMGD) is a new integrated platform that archives the mitochondrial genome sequences from 25,747 hexapod species, including 112 completely sequenced and 20 nearly completed genomes and 113,985 partially sequenced mitochondrial genomes. The Species-driven User Interface (SUI) of IMGD supports data retrieval and diverse analyses at multi-taxon levels. The Phyloviewer implemented in IMGD provides three methods for drawing phylogenetic trees and displays the resulting trees on the web. The SNP database incorporated to IMGD presents the distribution of SNPs and INDELs in the mitochondrial genomes of multiple isolates within eight species. A newly developed comparative SNU Genome Browser supports the graphical presentation and interactive interface for the identified SNPs/INDELs.
Conclusion: The IMGD provides a solid foundation for the comparative mitochondrial genomics and phylogenetics of insects. All data and functions described here are available at the web site (http://www.imgd.org/).
Figures



References
-
- Rawlings TA, Collins TM, Bieler R. A major mitochondrial gene rearrangement among closely related species. Mol Biol Evol. 2001;18:1604–1609. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

