Enrichment constrained time-dependent clustering analysis for finding meaningful temporal transcription modules
- PMID: 19351618
- PMCID: PMC2687989
- DOI: 10.1093/bioinformatics/btp235
Enrichment constrained time-dependent clustering analysis for finding meaningful temporal transcription modules
Abstract
Motivation: Clustering is a popular data exploration technique widely used in microarray data analysis. When dealing with time-series data, most conventional clustering algorithms, however, either use one-way clustering methods, which fail to consider the heterogeneity of temporary domain, or use two-way clustering methods that do not take into account the time dependency between samples, thus producing less informative results. Furthermore, enrichment analysis is often performed independent of and after clustering and such practice, though capable of revealing biological significant clusters, cannot guide the clustering to produce biologically significant result.
Result: We present a new enrichment constrained framework (ECF) coupled with a time-dependent iterative signature algorithm (TDISA), which, by applying a sliding time window to incorporate the time dependency of samples and imposing an enrichment constraint to parameters of clustering, allows supervised identification of temporal transcription modules (TTMs) that are biologically meaningful. Rigorous mathematical definitions of TTM as well as the enrichment constraint framework are also provided that serve as objective functions for retrieving biologically significant modules. We applied the enrichment constrained time-dependent iterative signature algorithm (ECTDISA) to human gene expression time-series data of Kaposi's sarcoma-associated herpesvirus (KSHV) infection of human primary endothelial cells; the result not only confirms known biological facts, but also reveals new insight into the molecular mechanism of KSHV infection.
Availability: Data and Matlab code are available at http://engineering.utsa.edu/ approximately yfhuang/ECTDISA.html.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
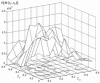






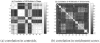
References
-
- Bergmann S, et al. Iterative signature algorithm for the analysis of large-scale gene expression data. Phys Rev. E Stat. Nonlin. Soft. Matter Phys. 2003;67:031902. - PubMed
-
- Bittner M, et al. Data analysis and integration: of steps and arrows. Nat. Genet. 1999;22:213–215. - PubMed
-
- Califano A, et al. Analysis of gene expression microarrays for phenotype classification. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:75–85. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

