Gene expression patterns in mismatch repair-deficient colorectal cancers highlight the potential therapeutic role of inhibitors of the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway
- PMID: 19351759
- PMCID: PMC3425357
- DOI: 10.1158/1078-0432.CCR-08-2432
Gene expression patterns in mismatch repair-deficient colorectal cancers highlight the potential therapeutic role of inhibitors of the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway
Abstract
Purpose: High-frequency microsatellite-instable (MSI-H) tumors account for approximately 15% of colorectal cancers. Therapeutic decisions for colorectal cancer are empirically based and currently do not emphasize molecular subclassification despite an increasing collection of gene expression information. Our objective was to identify low molecular weight compounds with preferential activity against MSI colorectal cancers using combined gene expression data sets.
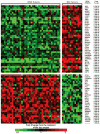
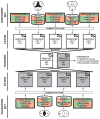
Experimental design: Three expression/query signatures (discovery data set) characterizing MSI-H colorectal cancer were matched with information derived from changes induced in cell lines by 164 compounds using the systems biology tool "Connectivity Map." A series of sequential filtering and ranking algorithms were used to select the candidate compounds. Compounds were validated using two additional expression/query signatures (validation data set). Cytotoxic, cell cycle, and apoptosis effects of validated compounds were evaluated in a panel of cell lines.
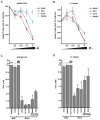
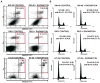
Results: Fourteen of the 164 compounds were validated as targeting MSI-H cell lines using the bioinformatics approach; rapamycin, LY-294002, 17-(allylamino)-17-demethoxygeldanamycin, and trichostatin A were the most robust candidate compounds. In vitro results showed that MSI-H cell lines due to hypermethylation of MLH1 are preferentially targeted by rapamycin (18.3 versus 4.4 mumol/L; P = 0.0824) and LY-294002 (15.02 versus 10.37 mumol/L; P = 0.0385) when compared with microsatellite-stable cells. Preferential activity was also observed in MSH2 and MSH6 mutant cells.
Conclusion: Our study shows that the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway is of special relevance in mismatch repair-deficient colorectal cancer. In addition, we show that amalgamation of gene expression information across studies provides a robust approach for selection of potential therapies corresponding to specific groups of patients.
Figures

References
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- SEER. US Mortality Age-Adjusted Rates, Total US 1990–1999: US National Center for Health Statistics. 2002.
-
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. - PubMed
-
- Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–7. - PubMed
-
- Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

