Strong evidence for lineage and sequence specificity of substitution rates and patterns in Drosophila
- PMID: 19351792
- PMCID: PMC2734147
- DOI: 10.1093/molbev/msp071
Strong evidence for lineage and sequence specificity of substitution rates and patterns in Drosophila
Abstract
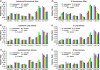
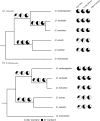
Rates of single nucleotide substitution in Drosophila are highly variable within the genome, and several examples illustrate that evolutionary rates differ among Drosophila species as well. Here, we use a maximum likelihood method to quantify lineage-specific substitutional patterns and apply this method to 4-fold degenerate synonymous sites and introns from more than 8,000 genes aligned in the Drosophila melanogaster group. We find that within species, different classes of sequence evolve at different rates, with long introns evolving most slowly and short introns evolving most rapidly. Relative rates of individual single nucleotide substitutions vary approximately 3-fold among lineages, yielding patterns of substitution that are comparatively less GC-biased in the melanogaster species complex relative to Drosophila yakuba and Drosophila erecta. These results are consistent with a model coupling a mutational shift toward reduced GC content, or a shift in mutation-selection balance, in the D. melanogaster species complex, with variation in selective constraint among different classes of DNA sequence. Finally, base composition of coding and intronic sequences is not at equilibrium with respect to substitutional patterns, which primarily reflects the slow rate of the substitutional process. These results thus support the view that mutational and/or selective processes are labile on an evolutionary timescale and that if the process is indeed selection driven, then the distribution of selective constraint is variable across the genome.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

