Applications of microarray technology to Acute Myelogenous Leukemia
- PMID: 19352456
- PMCID: PMC2664704
- DOI: 10.4137/cin.s1015
Applications of microarray technology to Acute Myelogenous Leukemia
Abstract
Microarray technology is a powerful tool, which has been applied to further the understanding of gene expression changes in disease. Array technology has been applied to the diagnosis and prognosis of Acute Myelogenous Leukemia (AML). Arrays have also been used extensively in elucidating the mechanism of and predicting therapeutic response in AML, as well as to further define the mechanism of AML pathogenesis. In this review, we discuss the major paradigms of gene expression array analysis, and provide insights into the use of software tools to annotate the array dataset and elucidate deregulated pathways and gene interaction networks. We present the application of gene expression array technology to questions in acute myelogenous leukemia; specifically, disease diagnosis, treatment and prognosis, and disease pathogenesis. Finally, we discuss several new and emerging array technologies, and how they can be further utilized to improve our understanding of AML.
Keywords: acute myelogenous leukemia; diagnostics; downstream genetic targets; gene expression profiling; prognosis; therapeutics.
Figures

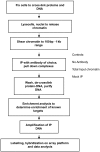
Similar articles
-
Large-scale gene network analysis reveals the significance of extracellular matrix pathway and homeobox genes in acute myeloid leukemia: an introduction to the Pigengene package and its applications.BMC Med Genomics. 2017 Mar 16;10(1):16. doi: 10.1186/s12920-017-0253-6. BMC Med Genomics. 2017. PMID: 28298217 Free PMC article.
-
Gene expression profiling in acute myeloid leukemia.J Clin Oncol. 2005 Sep 10;23(26):6296-305. doi: 10.1200/JCO.2005.05.020. J Clin Oncol. 2005. PMID: 16155012 Review.
-
Stratification of acute myeloid leukemia based on gene expression profiles.Int J Hematol. 2004 Dec;80(5):389-94. doi: 10.1532/ijh97.04111. Int J Hematol. 2004. PMID: 15646648 Review.
-
Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR.Int J Oncol. 2018 Mar;52(3):656-678. doi: 10.3892/ijo.2017.4233. Epub 2017 Dec 28. Int J Oncol. 2018. PMID: 29286103 Free PMC article. Clinical Trial.
-
Proteomics in acute myelogenous leukaemia (AML): methodological strategies and identification of protein targets for novel antileukaemic therapy.Curr Drug Targets. 2005 Sep;6(6):631-46. doi: 10.2174/1389450054863671. Curr Drug Targets. 2005. PMID: 16178797 Review.
Cited by
-
Deep Transcriptome Sequencing of Pediatric Acute Myeloid Leukemia Patients at Diagnosis, Remission and Relapse: Experience in 3 Malaysian Children in a Single Center Study.Front Genet. 2020 Feb 27;11:66. doi: 10.3389/fgene.2020.00066. eCollection 2020. Front Genet. 2020. PMID: 32174960 Free PMC article. No abstract available.
-
Breakthroughs within reach.Nat Med. 2009 May;15(5):476-9. doi: 10.1038/nm0509-476. Nat Med. 2009. PMID: 19424199 No abstract available.
-
Gene expression profiling unveils the temporal dynamics of CIGB-300-regulated transcriptome in AML cell lines.BMC Genomics. 2023 Jul 4;24(1):373. doi: 10.1186/s12864-023-09472-5. BMC Genomics. 2023. PMID: 37400761 Free PMC article.
References
-
- Agrawal S, et al. The C/EBPdelta tumor suppressor is silenced by hypermethylation in acute myeloid leukemia. Blood. 2007;109:3895–905. - PubMed
-
- Alcalay M, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. - PubMed
-
- Alevizos I, et al. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20:6196–204. - PubMed
-
- Aster JC. Diseases of white blood cells, lymph nodes, spleen and thymus. In: Kumar V, Abbas AK, Fausto N, editors. Robbins and Cotran: Pathologic Basis of Disease . Philadelphia: Elsevier Saunders; 2005. Ch(14)
LinkOut - more resources
Full Text Sources

