Direct quantitation of peptide mixtures without standards using clusters formed by electrospray ionization mass spectrometry
- PMID: 19354265
- PMCID: PMC4251781
- DOI: 10.1021/ac900294r
Direct quantitation of peptide mixtures without standards using clusters formed by electrospray ionization mass spectrometry
Abstract
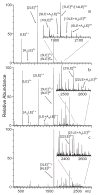
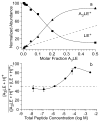
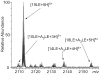
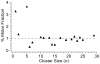
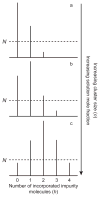
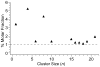
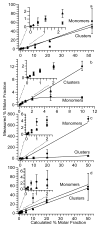
In electrospray ionization mass spectrometry, ion abundances depend on a number of different factors, including analyte surface activity, competition between analytes for charge, analyte concentration, as well as instrumental factors, including mass-dependent ion transmission and detection. Here, a novel method for obtaining quantitative information about solution-phase concentrations of peptide mixtures is described and demonstrated for five different peptide mixtures with relative concentrations ranging from 0.05% to 50%. In this method, the abundances of large clusters containing anywhere from 0 to 13 impurity molecules are measured and directly related to the relative solution-phase concentration of the peptides. For clusters containing approximately 15 or more peptides, the composition of the clusters approaches the statistical value indicating that these clusters are formed nonspecifically and that any differences in ion detection or ionization efficiency are negligible at these large cluster sizes. This method is accurate to within approximately 20% or better, even when the relative ion intensities of the protonated monomers can differ by over an order of magnitude compared to their solution-phase concentrations. Although less accurate than other quantitation methods that employ internal standards, this method does have the key advantages of speed, simplicity, and the ability to quantitate components in solution even when the identities of the components are unknown.
Figures

Similar articles
-
Simultaneous quantitation of amino acid mixtures using clustering agents.J Am Soc Mass Spectrom. 2011 Apr;22(4):624-32. doi: 10.1007/s13361-011-0081-4. Epub 2011 Feb 25. J Am Soc Mass Spectrom. 2011. PMID: 21472601 Free PMC article.
-
Standard-free quantitation of mixtures using clusters formed by electrospray mass spectrometry.Anal Chem. 2009 Oct 15;81(20):8434-40. doi: 10.1021/ac901405w. Anal Chem. 2009. PMID: 19754104 Free PMC article.
-
Comparison of the effects of ionization mechanism, analyte concentration, and ion "cool-times" on the internal energies of peptide ions produced by electrospray and atmospheric pressure matrix-assisted laser desorption ionization.J Am Soc Mass Spectrom. 2005 May;16(5):743-51. doi: 10.1016/j.jasms.2005.01.018. J Am Soc Mass Spectrom. 2005. PMID: 15862775
-
Enhancing detection and characterization of lipids using charge manipulation in electrospray ionization-tandem mass spectrometry.Chem Phys Lipids. 2020 Oct;232:104970. doi: 10.1016/j.chemphyslip.2020.104970. Epub 2020 Sep 3. Chem Phys Lipids. 2020. PMID: 32890498 Free PMC article. Review.
-
Leucine enkephalin--a mass spectrometry standard.Mass Spectrom Rev. 2011 Mar-Apr;30(2):298-320. doi: 10.1002/mas.20279. Epub 2010 Jul 28. Mass Spectrom Rev. 2011. PMID: 20669325 Review.
Cited by
-
Native mass spectrometry-based metabolomics identifies metal-binding compounds.Nat Chem. 2022 Jan;14(1):100-109. doi: 10.1038/s41557-021-00803-1. Epub 2021 Nov 18. Nat Chem. 2022. PMID: 34795435 Free PMC article.
-
Simultaneous quantitation of amino acid mixtures using clustering agents.J Am Soc Mass Spectrom. 2011 Apr;22(4):624-32. doi: 10.1007/s13361-011-0081-4. Epub 2011 Feb 25. J Am Soc Mass Spectrom. 2011. PMID: 21472601 Free PMC article.
-
Standard-free quantitation of mixtures using clusters formed by electrospray mass spectrometry.Anal Chem. 2009 Oct 15;81(20):8434-40. doi: 10.1021/ac901405w. Anal Chem. 2009. PMID: 19754104 Free PMC article.
-
Direct standard-free quantitation of Tamiflu and other pharmaceutical tablets using clustering agents with electrospray ionization mass spectrometry.Anal Chem. 2010 Feb 15;82(4):1179-82. doi: 10.1021/ac902277d. Anal Chem. 2010. PMID: 20092258 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

