Mapping protein-protein interactions by localized oxidation: consequences of the reach of hydroxyl radical
- PMID: 19354299
- PMCID: PMC2713361
- DOI: 10.1021/bi900273j
Mapping protein-protein interactions by localized oxidation: consequences of the reach of hydroxyl radical
Abstract
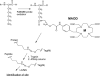
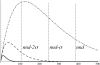
Hydroxyl radicals generated from a variety of methods, including not only synchrotron radiation but also Fenton reactions involving chelated iron, have become an accepted macromolecular footprinting tool. Hydroxyl radicals react with proteins via multiple mechanisms that lead to both polypeptide backbone cleavage events and side chain modifications (e.g., hydroxylation and carbonyl formation). The use of site-specifically tethered iron chelates can reveal protein-protein interactions, but the interpretation of such experiments will be strengthened by improving our understanding of how hydroxyl radicals produced at a point on a protein react with other protein sites. We have developed methods for monitoring carbonyl formation on proteins as a function of distance from a hydroxyl generator, iron-(S)-1-[p-(bromoacetamido)benzyl]EDTA (FeBABE), conjugated to an engineered cysteine residue. After activation of the chelated iron with ascorbate and peroxide produces new protein carbonyl groups, their positions can be identified using element-coded affinity tagging (ECAT), with carbonyl-specific tags {e.g., rare earth chelates of (S)-2-[4-(2-aminooxy)acetamidobenzyl]-1,4,7,10-tetraazacyclododecane-N,N',N'',N'''-tetraacetic acid (AOD)} that allow for affinity purification, identification, and relative quantitation of oxidation sites using mass spectrometry. Intraprotein oxidation of single-cysteine mutants of Escherichia coli sigma(70) by tethered FeBABE was used to calibrate the reach of hydroxyl radical by comparison to the crystal structure; the application to protein-protein interactions was demonstrated using the same sigma(70) FeBABE conjugates in complexes with the RNA polymerase core enzyme. The results provide fundamental information for interpreting protein footprinting experiments in other systems.
Figures




Similar articles
-
Mapping protein-protein interactions with a library of tethered cutting reagents: the binding site of sigma 70 on Escherichia coli RNA polymerase.Biochemistry. 1999 Apr 6;38(14):4259-65. doi: 10.1021/bi983016z. Biochemistry. 1999. PMID: 10194343
-
Mapping RNA-protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe.J Mol Biol. 2000 Feb 11;296(1):19-31. doi: 10.1006/jmbi.1999.3443. J Mol Biol. 2000. PMID: 10656815
-
Organization of open complexes at Escherichia coli promoters. Location of promoter DNA sites close to region 2.5 of the sigma70 subunit of RNA polymerase.J Biol Chem. 1999 Jan 22;274(4):2263-70. doi: 10.1074/jbc.274.4.2263. J Biol Chem. 1999. PMID: 9890989
-
Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization.Acc Chem Res. 2018 Mar 20;51(3):736-744. doi: 10.1021/acs.accounts.7b00593. Epub 2018 Feb 16. Acc Chem Res. 2018. PMID: 29450991 Free PMC article. Review.
-
[Free oxygen radiacals and kidney diseases--part I].Med Pregl. 2000 Sep-Oct;53(9-10):463-74. Med Pregl. 2000. PMID: 11320727 Review. Croatian.
Cited by
-
Amyloid fiber formation in human γD-Crystallin induced by UV-B photodamage.Biochemistry. 2013 Sep 10;52(36):6169-81. doi: 10.1021/bi4008353. Epub 2013 Aug 29. Biochemistry. 2013. PMID: 23957864 Free PMC article.
-
Protein oxidation of fucose environments (POFE) reveals fucose-protein interactions.Chem Sci. 2024 Mar 11;15(14):5256-5267. doi: 10.1039/d3sc06432h. eCollection 2024 Apr 3. Chem Sci. 2024. PMID: 38577366 Free PMC article.
-
Bordetella pertussis fim3 gene regulation by BvgA: phosphorylation controls the formation of inactive vs. active transcription complexes.Proc Natl Acad Sci U S A. 2015 Feb 10;112(6):E526-35. doi: 10.1073/pnas.1421045112. Epub 2015 Jan 26. Proc Natl Acad Sci U S A. 2015. PMID: 25624471 Free PMC article.
-
The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase.J Mol Biol. 2010 Sep 3;401(5):708-25. doi: 10.1016/j.jmb.2010.06.036. Epub 2010 Jun 25. J Mol Biol. 2010. PMID: 20600118 Free PMC article.
-
Modeling of the DNA-binding site of yeast Pms1 by mass spectrometry.DNA Repair (Amst). 2011 May 5;10(5):454-65. doi: 10.1016/j.dnarep.2011.01.010. Epub 2011 Feb 26. DNA Repair (Amst). 2011. PMID: 21354867 Free PMC article.
References
-
- Xu G, Chance MR. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev. 2007;107:3514–3543. - PubMed
-
- Chen H-T, Hahn S. Mapping the Location of TFIIB within the RNA Polymerase II Transcription Preinitiation Complex: A Model for the Structure of the PIC. Cell. 2004;119:169–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

