Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease
- PMID: 19357089
- PMCID: PMC2685627
- DOI: 10.1105/tpc.107.055228
Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease
Abstract
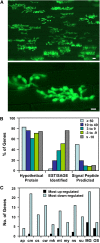
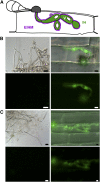
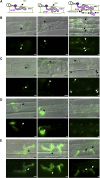
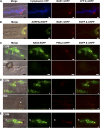
Biotrophic invasive hyphae (IH) of the blast fungus Magnaporthe oryzae secrete effectors to alter host defenses and cellular processes as they successively invade living rice (Oryza sativa) cells. However, few blast effectors have been identified. Indeed, understanding fungal and rice genes contributing to biotrophic invasion has been difficult because so few plant cells have encountered IH at the earliest infection stages. We developed a robust procedure for isolating infected-rice sheath RNAs in which approximately 20% of the RNA originated from IH in first-invaded cells. We analyzed these IH RNAs relative to control mycelial RNAs using M. oryzae oligoarrays. With a 10-fold differential expression threshold, we identified known effector PWL2 and 58 candidate effectors. Four of these candidates were confirmed to be fungal biotrophy-associated secreted (BAS) proteins. Fluorescently labeled BAS proteins were secreted into rice cells in distinct patterns in compatible, but not in incompatible, interactions. BAS1 and BAS2 proteins preferentially accumulated in biotrophic interfacial complexes along with known avirulence effectors, BAS3 showed additional localization near cell wall crossing points, and BAS4 uniformly outlined growing IH. Analysis of the same infected-tissue RNAs with rice oligoarrays identified putative effector-induced rice susceptibility genes, which are highly enriched for sensor-transduction components rather than typically identified defense response genes.
Figures
References
-
- Avilla-Adams, C., and Köller, W. (2002). Disruption of the alternative oxidase gene in Magnaporthe grisea and its impact on host infection. Mol. Plant Microbe Interact. 15 493–500. - PubMed
-
- Berruyer, R., Poussier, S., Kankanala, P., Mosquera, G., and Valent, B. (2006). Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology 96 346–355. - PubMed
-
- Bourett, T.M., Sweigard, J.A., Czymmek, K.J., Carroll, A., and Howard, R.J. (2002). Reef coral fluorescent proteins for visulaizing fungal pathogens. Fungal Genet. Biol. 37 211–220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

