Shared and group-specific features of the rotavirus RNA polymerase reveal potential determinants of gene reassortment restriction
- PMID: 19357162
- PMCID: PMC2687386
- DOI: 10.1128/JVI.00409-09
Shared and group-specific features of the rotavirus RNA polymerase reveal potential determinants of gene reassortment restriction
Abstract
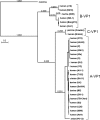
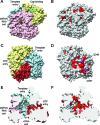
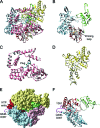
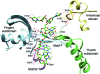
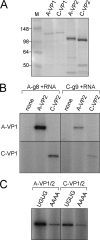
Rotaviruses (RVs) are nonenveloped, 11-segmented, double-stranded RNA viruses that are major pathogens associated with acute gastroenteritis. Group A, B, and C RVs have been isolated from humans; however, intergroup gene reassortment does not occur for reasons that remain unclear. This restriction might reflect the failure of the viral RNA-dependent RNA polymerase (RdRp; VP1) to recognize and replicate the RNA of a different group. To address this possibility, we contrasted the sequences, structures, and functions of RdRps belonging to RV groups A, B, and C (A-VP1, B-VP1, and C-VP1, respectively). We found that conserved amino acid residues are located within the hollow center of VP1 near the active site, whereas variable, group-specific residues are mostly surface exposed. By creating a three-dimensional homology model of C-VP1 with the A-VP1 crystallographic data, we provide evidence that these RV RdRps are nearly identical in their tertiary folds and that they have the same RNA template recognition mechanism that differs from that of B-VP1. Consistent with the structural data, recombinant A-VP1 and C-VP1 are capable of replicating one another's RNA templates in vitro. Nonetheless, the activity of both RdRps is strictly dependent upon the presence of cognate RV core shell protein A-VP2 or C-VP2, respectively. Together, the results of this study provide unprecedented insight into the structure and function of RV RdRps and support the notion that VP1 interactions may influence the emergence of reassortant viral strains.
Figures
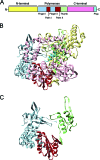

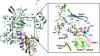
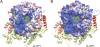
Similar articles
-
Group A Rotavirus VP1 Polymerase and VP2 Core Shell Proteins: Intergenotypic Sequence Variation and In Vitro Functional Compatibility.J Virol. 2019 Jan 4;93(2):e01642-18. doi: 10.1128/JVI.01642-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30355692 Free PMC article.
-
In Vitro Double-Stranded RNA Synthesis by Rotavirus Polymerase Mutants with Lesions at Core Shell Contact Sites.J Virol. 2019 Sep 30;93(20):e01049-19. doi: 10.1128/JVI.01049-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31341048 Free PMC article.
-
Rotavirus VP2 core shell regions critical for viral polymerase activation.J Virol. 2011 Apr;85(7):3095-105. doi: 10.1128/JVI.02360-10. Epub 2011 Jan 19. J Virol. 2011. PMID: 21248043 Free PMC article.
-
Rotavirus RNA replication and gene expression.Novartis Found Symp. 2001;238:64-77; discussion 77-81. doi: 10.1002/0470846534.ch5. Novartis Found Symp. 2001. PMID: 11444036 Review.
-
Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae.Virus Res. 2004 Apr;101(1):57-66. doi: 10.1016/j.virusres.2003.12.006. Virus Res. 2004. PMID: 15010217 Review.
Cited by
-
Residues of the rotavirus RNA-dependent RNA polymerase template entry tunnel that mediate RNA recognition and genome replication.J Virol. 2011 Mar;85(5):1958-69. doi: 10.1128/JVI.01689-10. Epub 2010 Dec 8. J Virol. 2011. PMID: 21147920 Free PMC article.
-
A Temperature-Sensitive Lesion in the N-Terminal Domain of the Rotavirus Polymerase Affects Its Intracellular Localization and Enzymatic Activity.J Virol. 2017 Mar 13;91(7):e00062-17. doi: 10.1128/JVI.00062-17. Print 2017 Apr 1. J Virol. 2017. PMID: 28100623 Free PMC article.
-
Genetic divergence of rotavirus nonstructural protein 4 results in distinct serogroup-specific viroporin activity and intracellular punctate structure morphologies.J Virol. 2012 May;86(9):4921-34. doi: 10.1128/JVI.06759-11. Epub 2012 Feb 22. J Virol. 2012. PMID: 22357281 Free PMC article.
-
The ins and outs of four-tunneled Reoviridae RNA-dependent RNA polymerases.Curr Opin Struct Biol. 2009 Dec;19(6):775-82. doi: 10.1016/j.sbi.2009.10.007. Epub 2009 Nov 14. Curr Opin Struct Biol. 2009. PMID: 19914820 Free PMC article. Review.
-
Group A Rotavirus VP1 Polymerase and VP2 Core Shell Proteins: Intergenotypic Sequence Variation and In Vitro Functional Compatibility.J Virol. 2019 Jan 4;93(2):e01642-18. doi: 10.1128/JVI.01642-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30355692 Free PMC article.
References
-
- Bremont, M., P. Juste-Lesage, D. Chabanne-Vautherot, A. Charpilienne, and J. Cohen. 1992. Sequences of the four larger proteins of a porcine group C rotavirus and comparison with the equivalent group A rotavirus proteins. Virology 186684-692. - PubMed
-
- Chen, Z., P. R. Lambden, J. Lau, E. O. Caul, and I. N. Clarke. 2002. Human group C rotavirus: completion of the genome sequence and gene coding assignments of a non-cultivatable rotavirus. Virus Res. 83179-187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

