A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function
- PMID: 19357306
- PMCID: PMC2678431
- DOI: 10.1073/pnas.0900833106
A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function
Abstract
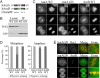
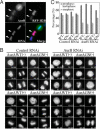
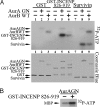
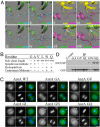
Aurora kinase-A and -B are key regulators of the cell cycle and tumorigenesis. It has remained a mystery why these 2 Aurora kinases, although highly similar in protein sequence and structure, are distinct in subcellular localization and function. Here, we report the striking finding that a single amino acid residue is responsible for these differences. We replaced the Gly-198 of Aurora-A with the equivalent residue Asn-142 of Aurora-B and found that in HeLa cells, Aurora-A(G198N) was recruited to the inner centromere in metaphase and the midzone in anaphase, reminiscent of the Aurora-B localization. Moreover, Aurora-A(G198N) compensated for the loss of Aurora-B in chromosome misalignment and cell premature exit from mitosis. Furthermore, Aurora-A(G198N) formed a complex with the Aurora-B partners, INCENP and Survivin, and its localization depended on this interaction. Aurora-A(G198N) phosphorylated the Aurora-B substrates INCENP and Survivin in vitro. Therefore, we propose that the presence of Gly or Asn at a single site assigns Aurora-A and -B to their respective partners and thus to their distinctive subcellular localizations and functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Taylor S, Peters JM. Polo and Aurora kinases: Lessons derived from chemical biology. Curr Opin Cell Biol. 2008;20:77–84. - PubMed
-
- Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. - PubMed
-
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. - PubMed
-
- Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. - PubMed
-
- Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

