Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity
- PMID: 19357332
- PMCID: PMC2712272
- DOI: 10.1152/jn.90941.2008
Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity
Erratum in
- J Neurophysiol. 2009 Sep;102(3):2008
Abstract
We consider the problem of extracting smooth, low-dimensional neural trajectories that summarize the activity recorded simultaneously from many neurons on individual experimental trials. Beyond the benefit of visualizing the high-dimensional, noisy spiking activity in a compact form, such trajectories can offer insight into the dynamics of the neural circuitry underlying the recorded activity. Current methods for extracting neural trajectories involve a two-stage process: the spike trains are first smoothed over time, then a static dimensionality-reduction technique is applied. We first describe extensions of the two-stage methods that allow the degree of smoothing to be chosen in a principled way and that account for spiking variability, which may vary both across neurons and across time. We then present a novel method for extracting neural trajectories-Gaussian-process factor analysis (GPFA)-which unifies the smoothing and dimensionality-reduction operations in a common probabilistic framework. We applied these methods to the activity of 61 neurons recorded simultaneously in macaque premotor and motor cortices during reach planning and execution. By adopting a goodness-of-fit metric that measures how well the activity of each neuron can be predicted by all other recorded neurons, we found that the proposed extensions improved the predictive ability of the two-stage methods. The predictive ability was further improved by going to GPFA. From the extracted trajectories, we directly observed a convergence in neural state during motor planning, an effect that was shown indirectly by previous studies. We then show how such methods can be a powerful tool for relating the spiking activity across a neural population to the subject's behavior on a single-trial basis. Finally, to assess how well the proposed methods characterize neural population activity when the underlying time course is known, we performed simulations that revealed that GPFA performed tens of percent better than the best two-stage method.
Figures

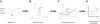


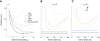






References
-
- Aksay E, Major G, Goldman MS, Baker R, Seung HS, Tank DW. History dependence of rate covariation between neurons during persistent activity in an oculomotor integrator. Cereb Cortex 13: 1173–1184, 2003. - PubMed
-
- Arieli A, Sterkin A, Grinvald A, Aertsen A. Dynamics of ongoing activity: explanation of the large variability in evoked cortical responses. Science 273: 1868–1871, 1996. - PubMed
-
- Bathellier B, Buhl DL, Accolla R, Carleton A. Dynamic ensemble odor coding in the mammalian olfactory bulb: sensory information at different timescales. Neuron 57: 586–598, 2008. - PubMed
-
- Batista AP, Santhanam G, Yu BM, Ryu SI, Afshar A, Shenoy KV. Reference frames for reach planning in macaque dorsal premotor cortex. J Neurophysiol 98: 966–983, 2007. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

