A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis
- PMID: 19359250
- PMCID: PMC2685685
- DOI: 10.1074/jbc.M109.004812
A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis
Abstract
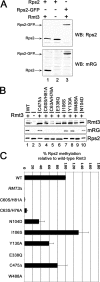
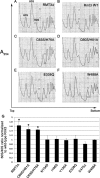
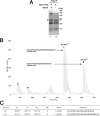
Schizosaccharomyces pombe Rmt3 is a member of the protein-arginine methyltransferase (PRMT) family and is the homolog of human PRMT3. We previously characterized Rmt3 as a ribosomal protein methyltransferase based on the identification of the 40 S Rps2 (ribosomal protein S2) as a substrate of Rmt3. RMT3-null cells produce nonmethylated Rps2 and show mis-regulation of the 40 S/60 S ribosomal subunit ratio due to a small subunit deficit. For this study, we have generated a series of RMT3 alleles that express various amino acid substitutions to characterize the functional domains of Rmt3 in Rps2 binding, Rps2 arginine methylation, and small ribosomal subunit production. Notably, catalytically inactive versions of Rmt3 restored the ribosomal subunit imbalance detected in RMT3-null cells. Consistent with a methyltransferase-independent function for Rmt3 in small ribosomal subunit production, the expression of an Rps2 variant in which the identified methylarginine residues were substituted with lysines showed normal levels of 40 S subunit. Importantly, substitutions within the zinc finger domain of Rmt3 that abolished Rps2 binding did not rescue the 40 S ribosomal subunit deficit of RMT3-null cells. Our findings suggest that the Rmt3-Rps2 interaction, rather than Rps2 methylation, is important for the function of Rmt3 in the regulation of small ribosomal subunit production.
Figures
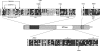


References
-
- Bedford, M. T., and Richard, S. (2005) Mol. Cell 18 263-272 - PubMed
-
- Bedford, M. T. (2007) J. Cell Sci. 120 4243-4246 - PubMed
-
- Gary, J. D., and Clarke, S. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61 65-131 - PubMed
-
- Kwak, Y. T., Guo, J., Prajapati, S., Park, K. J., Surabhi, R. M., Miller, B., Gehrig, P., and Gaynor, R. B. (2003) Mol. Cell 11 1055-1066 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

