Probing the mitotic history and developmental stage of hematopoietic cells using single telomere length analysis (STELA)
- PMID: 19359409
- PMCID: PMC2700317
- DOI: 10.1182/blood-2009-01-198374
Probing the mitotic history and developmental stage of hematopoietic cells using single telomere length analysis (STELA)
Abstract
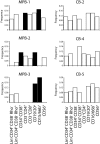
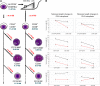
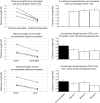
In most human somatic cells, telomeres shorten as a function of DNA replication. Telomere length is therefore an indirect measure of the replicative history of cells. We measured the telomere lengths at XpYp chromosomes in purified human hematopoietic populations enriched for stem cells (Lin(-)CD34(+)CD38(-)Rho(-)) and successively more mature cells. The average telomere length showed expected length changes, pointing to the utility of this method for classifying novel differentiation markers. Interestingly, the frequency of abruptly shortened telomeres increased in terminally differentiated adult populations, suggesting that damage to telomeric DNA occurs or is not repaired upon hematopoietic differentiation. When Lin(-)CD34(+)CD38(-)Rho(-) cord blood cells were transplanted into immunodeficient mice, the telomeres of the most primitive regenerated human hematopoietic cells lost approximately 3 kb, indicative of more than 30 cell divisions. Further losses in differentiating cells were similar to those observed in pretransplantation cell populations. These results indicate extensive self-renewal divisions of human hematopoietic stem cells are the primary cause of telomere erosion upon transplantation rather than added cell divisions in downstream progenitors.
Figures
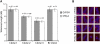


References
-
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. - PubMed
-
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. - PubMed
-
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. - PubMed
-
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

