Coding-sequence determinants of gene expression in Escherichia coli
- PMID: 19359587
- PMCID: PMC3902468
- DOI: 10.1126/science.1170160
Coding-sequence determinants of gene expression in Escherichia coli
Abstract
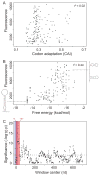
Synonymous mutations do not alter the encoded protein, but they can influence gene expression. To investigate how, we engineered a synthetic library of 154 genes that varied randomly at synonymous sites, but all encoded the same green fluorescent protein (GFP). When expressed in Escherichia coli, GFP protein levels varied 250-fold across the library. GFP messenger RNA (mRNA) levels, mRNA degradation patterns, and bacterial growth rates also varied, but codon bias did not correlate with gene expression. Rather, the stability of mRNA folding near the ribosomal binding site explained more than half the variation in protein levels. In our analysis, mRNA folding and associated rates of translation initiation play a predominant role in shaping expression levels of individual genes, whereas codon bias influences global translation efficiency and cellular fitness.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

