Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos
- PMID: 19360017
- PMCID: PMC2780369
- DOI: 10.1038/nprot.2009.55
Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos
Abstract
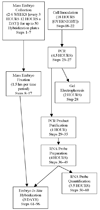
We describe a high-throughput protocol for RNA in situ hybridization (ISH) to Drosophila embryos in a 96-well format. cDNA or genomic DNA templates are amplified by PCR and then digoxigenin-labeled ribonucleotides are incorporated into antisense RNA probes by in vitro transcription. The quality of each probe is evaluated before ISH using a RNA probe quantification (dot blot) assay. RNA probes are hybridized to fixed, mixed-staged Drosophila embryos in 96-well plates. The resulting stained embryos can be examined and photographed immediately or stored at 4 degrees C for later analysis. Starting with fixed, staged embryos, the protocol takes 6 d from probe template production through hybridization. Preparation of fixed embryos requires a minimum of 2 weeks to collect embryos representing all stages. The method has been used to determine the expression patterns of over 6,000 genes throughout embryogenesis.
Figures



References
-
- Jones KW, Robertson FW. Localisation of reiterated nucleotide sequences in Drosophila and mouse by in situ hybridisation of complementary RNA. Chromosoma. 1970;31:331–345. - PubMed
-
- Harrison PR, Conkie D, Paul J, Jones K. Localisation of cellular globin messenger RNA by in situ hybridisation to complementary DNA. FEBS Lett. 1973;32:109–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

