The ups and downs of p53: understanding protein dynamics in single cells
- PMID: 19360021
- PMCID: PMC2892289
- DOI: 10.1038/nrc2604
The ups and downs of p53: understanding protein dynamics in single cells
Abstract
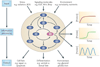
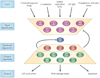
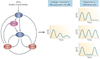
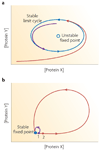
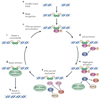
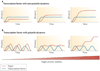
Cells living in a complex environment must constantly detect, process and appropriately respond to changing signals. Therefore, all cellular information processing is dynamic in nature. As a consequence, understanding the process of signal transduction often requires detailed quantitative analysis of dynamic behaviours. Here, we focus on the oscillatory dynamics of the tumour suppressor protein p53 as a model for studying protein dynamics in single cells to better understand its regulation and function.
Figures
Similar articles
-
[Telomere maintenance by DNA damage response machinery].Seikagaku. 2010 Dec;82(12):1145-50. Seikagaku. 2010. PMID: 21348272 Review. Japanese. No abstract available.
-
[P53-independent apoptosis through a novel ATM/IKK-alpha/p73-mediated apoptotic pathway].Seikagaku. 2008 May;80(5):409-13. Seikagaku. 2008. PMID: 18575226 Review. Japanese. No abstract available.
-
[Activation of p53 protein after DNA damages].Tanpakushitsu Kakusan Koso. 2001 Jun;46(8 Suppl):1188-93. Tanpakushitsu Kakusan Koso. 2001. PMID: 11436309 Review. Japanese. No abstract available.
-
Cell fate decision mediated by p53 pulses.Proc Natl Acad Sci U S A. 2009 Jul 28;106(30):12245-50. doi: 10.1073/pnas.0813088106. Epub 2009 Jul 15. Proc Natl Acad Sci U S A. 2009. PMID: 19617533 Free PMC article.
-
The impact of a negligent G2/M checkpoint on genomic instability and cancer induction.Nat Rev Cancer. 2007 Nov;7(11):861-9. doi: 10.1038/nrc2248. Nat Rev Cancer. 2007. PMID: 17943134 Review.
Cited by
-
Clinicopathologic Correlations of E-cadherin and Prrx-1 Expression Loss in Hepatocellular Carcinoma.J Pathol Transl Med. 2016 Sep;50(5):327-36. doi: 10.4132/jptm.2016.06.22. Epub 2016 Aug 31. J Pathol Transl Med. 2016. PMID: 27580127 Free PMC article.
-
Misuse of the Michaelis-Menten rate law for protein interaction networks and its remedy.PLoS Comput Biol. 2020 Oct 22;16(10):e1008258. doi: 10.1371/journal.pcbi.1008258. eCollection 2020 Oct. PLoS Comput Biol. 2020. PMID: 33090989 Free PMC article. Review.
-
Toxicity testing in the 21 century: defining new risk assessment approaches based on perturbation of intracellular toxicity pathways.PLoS One. 2011;6(6):e20887. doi: 10.1371/journal.pone.0020887. Epub 2011 Jun 20. PLoS One. 2011. PMID: 21701582 Free PMC article. Review.
-
Transcription factor oscillations in neural stem cells: Implications for accurate control of gene expression.Neurogenesis (Austin). 2017 Feb 10;4(1):e1262934. doi: 10.1080/23262133.2016.1262934. eCollection 2017. Neurogenesis (Austin). 2017. PMID: 28321433 Free PMC article. Review.
-
Can case study approaches speed implementation of the NRC report: "Toxicity Testing in the 21st Century: A Vision and a Strategy?".ALTEX. 2011;28(3):175-82. doi: 10.14573/altex.2011.3.175. ALTEX. 2011. PMID: 21993955 Free PMC article.
References
-
- Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. - PubMed
-
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. - PubMed
-
- Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. - PubMed
-
- Nelson DE, See V, Nelson G, White MR. Oscillations in transcription factor dynamics: a new way to control gene expression. Biochem Soc Trans. 2004;32:1090–1092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

