The ups and downs of p53: understanding protein dynamics in single cells
- PMID: 19360021
- PMCID: PMC2892289
- DOI: 10.1038/nrc2604
The ups and downs of p53: understanding protein dynamics in single cells
Abstract
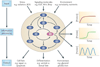
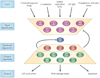
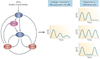
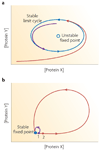
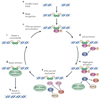
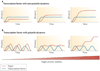
Cells living in a complex environment must constantly detect, process and appropriately respond to changing signals. Therefore, all cellular information processing is dynamic in nature. As a consequence, understanding the process of signal transduction often requires detailed quantitative analysis of dynamic behaviours. Here, we focus on the oscillatory dynamics of the tumour suppressor protein p53 as a model for studying protein dynamics in single cells to better understand its regulation and function.
Figures
References
-
- Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. - PubMed
-
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. - PubMed
-
- Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. - PubMed
-
- Nelson DE, See V, Nelson G, White MR. Oscillations in transcription factor dynamics: a new way to control gene expression. Biochem Soc Trans. 2004;32:1090–1092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

