Statistical comparison framework and visualization scheme for ranking-based algorithms in high-throughput genome-wide studies
- PMID: 19361328
- PMCID: PMC3148127
- DOI: 10.1089/cmb.2008.0151
Statistical comparison framework and visualization scheme for ranking-based algorithms in high-throughput genome-wide studies
Abstract
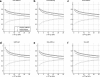
As a first step in analyzing high-throughput data in genome-wide studies, several algorithms are available to identify and prioritize candidates lists for downstream fine-mapping. The prioritized candidates could be differentially expressed genes, aberrations in comparative genomics hybridization studies, or single nucleotide polymorphisms (SNPs) in association studies. Different analysis algorithms are subject to various experimental artifacts and analytical features that lead to different candidate lists. However, little research has been carried out to theoretically quantify the consensus between different candidate lists and to compare the study specific accuracy of the analytical methods based on a known reference candidate list. Within the context of genome-wide studies, we propose a generic mathematical framework to statistically compare ranked lists of candidates from different algorithms with each other or, if available, with a reference candidate list. To cope with the growing need for intuitive visualization of high-throughput data in genome-wide studies, we describe a complementary customizable visualization tool. As a case study, we demonstrate application of our framework to the comparison and visualization of candidate lists generated in a DNA-pooling based genome-wide association study of CEPH data in the HapMap project, where prior knowledge from individual genotyping can be used to generate a true reference candidate list. The results provide a theoretical basis to compare the accuracy of various methods and to identify redundant methods, thus providing guidance for selecting the most suitable analysis method in genome-wide studies.
Figures


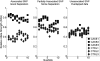
References
-
- Azuaje F. A cluster validity framework for genome expression data. Bioinformatics. 2002;18:319–320. - PubMed
-
- Brun M. Sima C. Hua J., et al. Model-based evaluation of clustering validation measures. Pattern Recogn. 2007;40:807–824.
-
- Dwork C. Kumar R. Naor M., et al. Rank aggregation methods for the web. WWW. 2001:613–622.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

