Massive comparative genomic analysis reveals convergent evolution of specialized bacteria
- PMID: 19361336
- PMCID: PMC2688493
- DOI: 10.1186/1745-6150-4-13
Massive comparative genomic analysis reveals convergent evolution of specialized bacteria
Abstract
Background: Genome size and gene content in bacteria are associated with their lifestyles. Obligate intracellular bacteria (i.e., mutualists and parasites) have small genomes that derived from larger free-living bacterial ancestors; however, the different steps of bacterial specialization from free-living to intracellular lifestyle have not been studied comprehensively. The growing number of available sequenced genomes makes it possible to perform a statistical comparative analysis of 317 genomes from bacteria with different lifestyles.
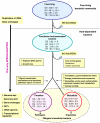
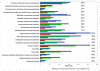
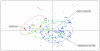
Results: Compared to free-living bacteria, host-dependent bacteria exhibit fewer rRNA genes, more split rRNA operons and fewer transcriptional regulators, linked to slower growth rates. We found a function-dependent and non-random loss of the same 100 orthologous genes in all obligate intracellular bacteria. Thus, we showed that obligate intracellular bacteria from different phyla are converging according to their lifestyle. Their specialization is an irreversible phenomenon characterized by translation modification and massive gene loss, including the loss of transcriptional regulators. Although both mutualists and parasites converge by genome reduction, these obligate intracellular bacteria have lost distinct sets of genes in the context of their specific host associations: mutualists have significantly more genes that enable nutrient provisioning whereas parasites have genes that encode Types II, IV, and VI secretion pathways.
Conclusion: Our findings suggest that gene loss, rather than acquisition of virulence factors, has been a driving force in the adaptation of parasites to eukaryotic cells. This comparative genomic analysis helps to explore the strategies by which obligate intracellular genomes specialize to particular host-associations and contributes to advance our knowledge about the mechanisms of bacterial evolution.
Figures


Similar articles
-
Ongoing Transposon-Mediated Genome Reduction in the Luminous Bacterial Symbionts of Deep-Sea Ceratioid Anglerfishes.mBio. 2018 Jun 26;9(3):e01033-18. doi: 10.1128/mBio.01033-18. mBio. 2018. PMID: 29946051 Free PMC article.
-
Genome evolution in bacterial endosymbionts of insects.Nat Rev Genet. 2002 Nov;3(11):850-61. doi: 10.1038/nrg931. Nat Rev Genet. 2002. PMID: 12415315 Review.
-
Paradoxical evolution of rickettsial genomes.Ticks Tick Borne Dis. 2019 Feb;10(2):462-469. doi: 10.1016/j.ttbdis.2018.11.007. Epub 2018 Nov 12. Ticks Tick Borne Dis. 2019. PMID: 30448253 Review.
-
Genome Analysis of Endomicrobium proavitum Suggests Loss and Gain of Relevant Functions during the Evolution of Intracellular Symbionts.Appl Environ Microbiol. 2017 Aug 17;83(17):e00656-17. doi: 10.1128/AEM.00656-17. Print 2017 Sep 1. Appl Environ Microbiol. 2017. PMID: 28646115 Free PMC article.
-
Genomics of epidemic pathogens.Clin Microbiol Infect. 2012 Mar;18(3):213-7. doi: 10.1111/j.1469-0691.2012.03781.x. Clin Microbiol Infect. 2012. PMID: 22369153 Review.
Cited by
-
A comparison of the endotoxin biosynthesis and protein oxidation pathways in the biogenesis of the outer membrane of Escherichia coli and Neisseria meningitidis.Front Cell Infect Microbiol. 2012 Dec 20;2:162. doi: 10.3389/fcimb.2012.00162. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23267440 Free PMC article. Review.
-
Reconstructing Gene Gains and Losses with BadiRate.Methods Mol Biol. 2022;2569:213-232. doi: 10.1007/978-1-0716-2691-7_10. Methods Mol Biol. 2022. PMID: 36083450 Review.
-
Comparative genomics reveals a novel genetic organization of the sad cluster in the sulfonamide-degrader 'Candidatus Leucobacter sulfamidivorax' strain GP.BMC Genomics. 2019 Nov 21;20(1):885. doi: 10.1186/s12864-019-6206-z. BMC Genomics. 2019. PMID: 31752666 Free PMC article.
-
Maturation of atypical ribosomal RNA precursors in Helicobacter pylori.Nucleic Acids Res. 2019 Jun 20;47(11):5906-5921. doi: 10.1093/nar/gkz258. Nucleic Acids Res. 2019. PMID: 31006803 Free PMC article.
-
Pangenome Evolution in Environmentally Transmitted Symbionts of Deep-Sea Mussels Is Governed by Vertical Inheritance.Genome Biol Evol. 2022 Jul 2;14(7):evac098. doi: 10.1093/gbe/evac098. Genome Biol Evol. 2022. PMID: 35731940 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

