EFICAz2: enzyme function inference by a combined approach enhanced by machine learning
- PMID: 19361344
- PMCID: PMC2670841
- DOI: 10.1186/1471-2105-10-107
EFICAz2: enzyme function inference by a combined approach enhanced by machine learning
Abstract
Background: We previously developed EFICAz, an enzyme function inference approach that combines predictions from non-completely overlapping component methods. Two of the four components in the original EFICAz are based on the detection of functionally discriminating residues (FDRs). FDRs distinguish between member of an enzyme family that are homofunctional (classified under the EC number of interest) or heterofunctional (annotated with another EC number or lacking enzymatic activity). Each of the two FDR-based components is associated to one of two specific kinds of enzyme families. EFICAz exhibits high precision performance, except when the maximal test to training sequence identity (MTTSI) is lower than 30%. To improve EFICAz's performance in this regime, we: i) increased the number of predictive components and ii) took advantage of consensual information from the different components to make the final EC number assignment.
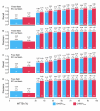
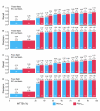
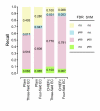
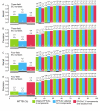
Results: We have developed two new EFICAz components, analogs to the two FDR-based components, where the discrimination between homo and heterofunctional members is based on the evaluation, via Support Vector Machine models, of all the aligned positions between the query sequence and the multiple sequence alignments associated to the enzyme families. Benchmark results indicate that: i) the new SVM-based components outperform their FDR-based counterparts, and ii) both SVM-based and FDR-based components generate unique predictions. We developed classification tree models to optimally combine the results from the six EFICAz components into a final EC number prediction. The new implementation of our approach, EFICAz2, exhibits a highly improved prediction precision at MTTSI < 30% compared to the original EFICAz, with only a slight decrease in prediction recall. A comparative analysis of enzyme function annotation of the human proteome by EFICAz2 and KEGG shows that: i) when both sources make EC number assignments for the same protein sequence, the assignments tend to be consistent and ii) EFICAz2 generates considerably more unique assignments than KEGG.
Conclusion: Performance benchmarks and the comparison with KEGG demonstrate that EFICAz2 is a powerful and precise tool for enzyme function annotation, with multiple applications in genome analysis and metabolic pathway reconstruction. The EFICAz2 web service is available at: http://cssb.biology.gatech.edu/skolnick/webservice/EFICAz2/index.html.
Figures
Similar articles
-
EFICAz: a comprehensive approach for accurate genome-scale enzyme function inference.Nucleic Acids Res. 2004 Dec 1;32(21):6226-39. doi: 10.1093/nar/gkh956. Print 2004. Nucleic Acids Res. 2004. PMID: 15576349 Free PMC article.
-
EFICAz2.5: application of a high-precision enzyme function predictor to 396 proteomes.Bioinformatics. 2012 Oct 15;28(20):2687-8. doi: 10.1093/bioinformatics/bts510. Epub 2012 Aug 24. Bioinformatics. 2012. PMID: 22923291 Free PMC article.
-
High precision multi-genome scale reannotation of enzyme function by EFICAz.BMC Genomics. 2006 Dec 13;7:315. doi: 10.1186/1471-2164-7-315. BMC Genomics. 2006. PMID: 17166279 Free PMC article.
-
A Survey for Predicting Enzyme Family Classes Using Machine Learning Methods.Curr Drug Targets. 2019;20(5):540-550. doi: 10.2174/1389450119666181002143355. Curr Drug Targets. 2019. PMID: 30277150 Review.
-
Enzyme function prediction with interpretable models.Methods Mol Biol. 2009;541:373-420. doi: 10.1007/978-1-59745-243-4_17. Methods Mol Biol. 2009. PMID: 19381539 Review.
Cited by
-
Horizontal gene transfer and genome evolution in Methanosarcina.BMC Evol Biol. 2015 Jun 5;15:102. doi: 10.1186/s12862-015-0393-2. BMC Evol Biol. 2015. PMID: 26044078 Free PMC article.
-
The use of evolutionary patterns in protein annotation.Curr Opin Struct Biol. 2012 Jun;22(3):316-25. doi: 10.1016/j.sbi.2012.05.001. Epub 2012 May 24. Curr Opin Struct Biol. 2012. PMID: 22633559 Free PMC article. Review.
-
Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis.Genome Biol. 2013 Jun 17;14(6):R60. doi: 10.1186/gb-2013-14-6-r60. Genome Biol. 2013. PMID: 23773524 Free PMC article.
-
Comparative genomics of cell envelope components in mycobacteria.PLoS One. 2011 May 6;6(5):e19280. doi: 10.1371/journal.pone.0019280. PLoS One. 2011. PMID: 21573108 Free PMC article.
-
Genomics of Loa loa, a Wolbachia-free filarial parasite of humans.Nat Genet. 2013 May;45(5):495-500. doi: 10.1038/ng.2585. Epub 2013 Mar 24. Nat Genet. 2013. PMID: 23525074 Free PMC article.
References
-
- Webb EC. Enzyme nomenclature 1992: recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes. San Diego: Published for the International Union of Biochemistry and Molecular Biology by Academic Press; 1992.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

