Insights into how RNase R degrades structured RNA: analysis of the nuclease domain
- PMID: 19361424
- PMCID: PMC2786168
- DOI: 10.1016/j.jmb.2009.01.068
Insights into how RNase R degrades structured RNA: analysis of the nuclease domain
Abstract
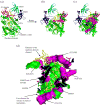
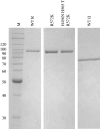
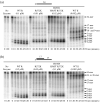
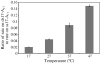
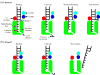
RNase R readily degrades highly structured RNA, whereas its paralogue, RNase II, is unable to do so. Furthermore, the nuclease domain of RNase R, devoid of all canonical RNA-binding domains, is sufficient for this activity. RNase R also binds RNA more tightly within its catalytic channel than does RNase II, which is thought to be important for its unique catalytic properties. To investigate this idea further, certain residues within the nuclease domain channel of RNase R were changed to those found in RNase II. Among the many examined, we identified one amino acid residue, R572, that has a significant role in the properties of RNase R. Conversion of this residue to lysine, as found in RNase II, results in weaker substrate binding within the nuclease domain channel, longer limit products, increased activity against a variety of substrates and a faster substrate on-rate. Most importantly, the mutant encounters difficulty in degrading structured RNA, pausing within a double-stranded region. Additional studies show that degradation of structured substrates is dependent upon temperature, suggesting a role for thermal breathing in the mechanism of action of RNase R. On the basis of these data, we propose a model in which tight binding within the nuclease domain allows RNase R to capitalize on the natural thermal breathing of an RNA duplex to degrade structured RNAs.
Figures

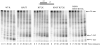

Similar articles
-
RNase R mutants elucidate the catalysis of structured RNA: RNA-binding domains select the RNAs targeted for degradation.Biochem J. 2009 Sep 25;423(2):291-301. doi: 10.1042/BJ20090839. Biochem J. 2009. PMID: 19630750
-
The roles of individual domains of RNase R in substrate binding and exoribonuclease activity. The nuclease domain is sufficient for digestion of structured RNA.J Biol Chem. 2009 Jan 2;284(1):486-494. doi: 10.1074/jbc.M806468200. Epub 2008 Nov 11. J Biol Chem. 2009. PMID: 19004832 Free PMC article.
-
How RNase R Degrades Structured RNA: ROLE OF THE HELICASE ACTIVITY AND THE S1 DOMAIN.J Biol Chem. 2016 Apr 8;291(15):7877-87. doi: 10.1074/jbc.M116.717991. Epub 2016 Feb 12. J Biol Chem. 2016. PMID: 26872969 Free PMC article.
-
RNase II: the finer details of the Modus operandi of a molecular killer.RNA Biol. 2010 May-Jun;7(3):276-81. doi: 10.4161/rna.7.3.11490. Epub 2010 May 9. RNA Biol. 2010. PMID: 20484980 Review.
-
Major 3'-5' Exoribonucleases in the Metabolism of Coding and Non-coding RNA.Prog Mol Biol Transl Sci. 2018;159:101-155. doi: 10.1016/bs.pmbts.2018.07.005. Epub 2018 Aug 25. Prog Mol Biol Transl Sci. 2018. PMID: 30340785 Review.
Cited by
-
Novel one-step mechanism for tRNA 3'-end maturation by the exoribonuclease RNase R of Mycoplasma genitalium.J Biol Chem. 2012 Jul 6;287(28):23427-33. doi: 10.1074/jbc.M111.324970. Epub 2012 May 17. J Biol Chem. 2012. PMID: 22605341 Free PMC article.
-
Through ancient rings thread programming strings.Structure. 2009 Nov 11;17(11):1429-31. doi: 10.1016/j.str.2009.10.006. Structure. 2009. PMID: 19913477 Free PMC article.
-
RNase II binds to RNase E and modulates its endoribonucleolytic activity in the cyanobacterium Anabaena PCC 7120.Nucleic Acids Res. 2020 Apr 17;48(7):3922-3934. doi: 10.1093/nar/gkaa092. Nucleic Acids Res. 2020. PMID: 32055835 Free PMC article.
-
Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains.Mol Microbiol. 2014 Oct;94(1):41-55. doi: 10.1111/mmi.12748. Epub 2014 Aug 21. Mol Microbiol. 2014. PMID: 25099370 Free PMC article.
-
Circular RNA discovery with emerging sequencing and deep learning technologies.Nat Genet. 2025 May;57(5):1089-1102. doi: 10.1038/s41588-025-02157-7. Epub 2025 Apr 17. Nat Genet. 2025. PMID: 40247051 Review.
References
-
- Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51:777–90. - PubMed
-
- Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

