The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation
- PMID: 19361448
- PMCID: PMC2677131
- DOI: 10.1016/j.jmb.2009.01.032
The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation
Abstract
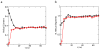
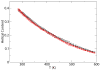
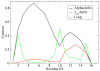
We have performed simulated tempering molecular dynamics simulations to study the thermodynamics of the headpiece of the Huntingtin (Htt) protein (N17(Htt)). With converged sampling, we found this peptide is highly helical, as previously proposed. Interestingly, this peptide is also found to adopt two different and seemingly stable states. The region from residue 4 (L) to residue 9 (K) has a strong helicity from our simulations, which is supported by experimental studies. However, contrary to what was initially proposed, we have found that simulations predict the most populated state as a two-helix bundle rather than a single straight helix, although a significant percentage of structures do still adopt a single linear helix. The fact that Htt aggregation is nucleation dependent infers the importance of a critical transition. It has been shown that N17(Htt) is involved in this rate-limiting step. In this study, we propose two possible mechanisms for this nucleating event stemming from the transition between two-helix bundle state and single-helix state for N17(Htt) and the experimentally observed interactions between the N17(Htt) and polyQ domains. More strikingly, an extensive hydrophobic surface area is found to be exposed to solvent in the dominant monomeric state of N17(Htt). We propose the most fundamental role played by N17(Htt) would be initializing the dimerization and pulling the polyQ chains into adequate spatial proximity for the nucleation event to proceed.
Figures

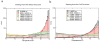



References
-
- Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, Watkins PC, Ottina K, Wallace MR, Sakaguchi AY, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–8. - PubMed
-
- Mac Donald ME, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–83. - PubMed
-
- Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman JJ, Chotai K, Connarty M, Crauford D, Curtis A, Curtis D, Davidson MJ, Differ AM, Dode C, Dodge A, Frontali M, Ranen NG, Stine OC, Sherr M, Abbott MH, Franz ML, Graham CA, Harper PS, Hedreen JC, Hayden MR, et al. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am J Hum Genet. 1996;59:16–22. - PMC - PubMed
-
- Sathasivam K, Amaechi I, Mangiarini L, Bates G. Identification of an HD patient with a (CAG)180 repeat expansion and the propagation of highly expanded CAG repeats in lambda phage. Hum Genet. 1997;99:692–5. - PubMed
-
- Masino L, Kelly G, Leonard K, Trottier Y, Pastore A. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett. 2002;513:267–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

