Genetic control of human brain transcript expression in Alzheimer disease
- PMID: 19361613
- PMCID: PMC2667989
- DOI: 10.1016/j.ajhg.2009.03.011
Genetic control of human brain transcript expression in Alzheimer disease
Abstract
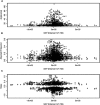
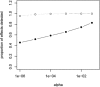
We recently surveyed the relationship between the human brain transcriptome and genome in a series of neuropathologically normal postmortem samples. We have now analyzed additional samples with a confirmed pathologic diagnosis of late-onset Alzheimer disease (LOAD; final n = 188 controls, 176 cases). Nine percent of the cortical transcripts that we analyzed had expression profiles correlated with their genotypes in the combined cohort, and approximately 5% of transcripts had SNP-transcript relationships that could distinguish LOAD samples. Two of these transcripts have been previously implicated in LOAD candidate-gene SNP-expression screens. This study shows how the relationship between common inherited genetic variants and brain transcript expression can be used in the study of human brain disorders. We suggest that studying the transcriptome as a quantitative endo-phenotype has greater power for discovering risk SNPs influencing expression than the use of discrete diagnostic categories such as presence or absence of disease.
Figures

References
-
- Schadt E.E. Exploiting naturally occurring DNA variation and molecular profiling data to dissect disease and drug response traits. Curr. Opin. Biotechnol. 2005;16:647–654. - PubMed
-
- Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. - PubMed
-
- Göring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. - PubMed
-
- Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. - PubMed
-
- Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

