U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome
- PMID: 19362093
- PMCID: PMC2682339
- DOI: 10.1016/j.jmb.2009.04.005
U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome
Abstract
Structures have been obtained for the complexes that tiamulin, homoharringtonine, and bruceantin form with the large ribosomal subunit of Haloarcula marismortui at resolutions ranging from 2.65 to 3.2 A. They show that all these inhibitors block protein synthesis by competing with the amino acid side chains of incoming aminoacyl-tRNAs for binding in the A-site cleft in the peptidyl-transferase center, which is universally conserved. In addition, these structures support the hypothesis that the species specificity exhibited by the A-site cleft inhibitors is determined by the interactions they make, or fail to make, with a single nucleotide, U2504 (Escherichia coli). In the ribosome, the position of U2504 is controlled by its interactions with neighboring nucleotides, whose identities vary among kingdoms.
Figures

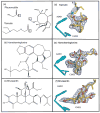
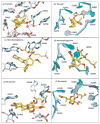
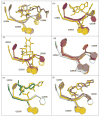


References
-
- Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. - PubMed
-
- Wilson DN, Harms JM, Nierhaus KH, Schlunzen F, Fucini P. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol Chem. 2005;386:1239–1252. - PubMed
-
- Vazquez D. Molecular Biology Biochemistry and Biopyhsics. Springer Verlag; New York: 1979. Inhibitors of Protein Biosynthesis. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

