An approximate likelihood for genetic data under a model with recombination and population splitting
- PMID: 19362099
- PMCID: PMC3108256
- DOI: 10.1016/j.tpb.2009.04.001
An approximate likelihood for genetic data under a model with recombination and population splitting
Abstract
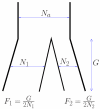
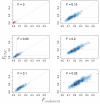
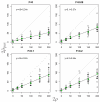
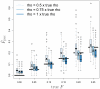
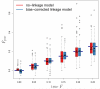
We describe a new approximate likelihood for population genetic data under a model in which a single ancestral population has split into two daughter populations. The approximate likelihood is based on the 'Product of Approximate Conditionals' likelihood and 'copying model' of Li and Stephens [Li, N., Stephens, M., 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165 (4), 2213-2233]. The approach developed here may be used for efficient approximate likelihood-based analyses of unlinked data. However our copying model also considers the effects of recombination. Hence, a more important application is to loosely-linked haplotype data, for which efficient statistical models explicitly featuring non-equilibrium population structure have so far been unavailable. Thus, in addition to the information in allele frequency differences about the timing of the population split, the method can also extract information from the lengths of haplotypes shared between the populations. There are a number of challenges posed by extracting such information, which makes parameter estimation difficult. We discuss how the approach could be extended to identify haplotypes introduced by migrants.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

