Toward a structural understanding of IRES RNA function
- PMID: 19362464
- PMCID: PMC2757110
- DOI: 10.1016/j.sbi.2009.03.005
Toward a structural understanding of IRES RNA function
Abstract
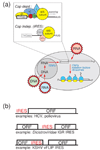
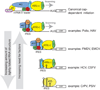
Protein synthesis of an RNA template can start by two different known mechanisms: cap-dependent translation initiation and cap-independent translation initiation. The latter is driven by RNA sequences called internal ribosome entry sites (IRESs) that are found in both viral RNAs and cellular mRNAs. The diverse mechanisms used by IRESs are reflected in their structural diversity, and this structural diversity challenges us to develop a cohesive model linking IRES function to structure. With more direct structural information available for the viral IRESs, data suggest an inverse correlation between the degree to which an IRES RNA can form a stable structure on its own and the number of factors that it requires to function. Lessons learned from the viral IRESs may help understand the cellular IRESs, although more structural data are needed before any strong links can be made.
Figures




Similar articles
-
Human IRES Atlas: an integrative platform for studying IRES-driven translational regulation in humans.Database (Oxford). 2021 May 3;2021:baab025. doi: 10.1093/database/baab025. Database (Oxford). 2021. PMID: 33942874 Free PMC article.
-
Ribosomal Protein L13 Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus in a Helicase DDX3-Dependent Manner.J Virol. 2020 Jan 6;94(2):e01679-19. doi: 10.1128/JVI.01679-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31619563 Free PMC article.
-
IRES-mediated cap-independent translation, a path leading to hidden proteome.J Mol Cell Biol. 2019 Oct 25;11(10):911-919. doi: 10.1093/jmcb/mjz091. J Mol Cell Biol. 2019. PMID: 31504667 Free PMC article. Review.
-
Unlike for cellular mRNAs and other viral internal ribosome entry sites (IRESs), the eIF3 subunit e is not required for the translational activity of the HCV IRES.J Biol Chem. 2020 Feb 14;295(7):1843-1856. doi: 10.1074/jbc.RA119.009502. Epub 2020 Jan 12. J Biol Chem. 2020. PMID: 31929110 Free PMC article.
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
Cited by
-
PSF: nuclear busy-body or nuclear facilitator?Wiley Interdiscip Rev RNA. 2015 Jul-Aug;6(4):351-67. doi: 10.1002/wrna.1280. Epub 2015 Apr 1. Wiley Interdiscip Rev RNA. 2015. PMID: 25832716 Free PMC article. Review.
-
La Autoantigen Induces Ribosome Binding Protein 1 (RRBP1) Expression through Internal Ribosome Entry Site (IRES)-Mediated Translation during Cellular Stress Condition.Int J Mol Sci. 2016 Jul 20;17(7):1174. doi: 10.3390/ijms17071174. Int J Mol Sci. 2016. PMID: 27447629 Free PMC article.
-
Cell type specificity and structural determinants of IRES activity from the 5' leaders of different HIV-1 transcripts.Nucleic Acids Res. 2013 Jul;41(13):6698-714. doi: 10.1093/nar/gkt358. Epub 2013 May 9. Nucleic Acids Res. 2013. PMID: 23661682 Free PMC article.
-
hnRNP K Is a Novel Internal Ribosomal Entry Site-Transacting Factor That Negatively Regulates Foot-and-Mouth Disease Virus Translation and Replication and Is Antagonized by Viral 3C Protease.J Virol. 2020 Aug 17;94(17):e00803-20. doi: 10.1128/JVI.00803-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581104 Free PMC article.
-
Production and Application of Multicistronic Constructs for Various Human Disease Therapies.Pharmaceutics. 2019 Nov 6;11(11):580. doi: 10.3390/pharmaceutics11110580. Pharmaceutics. 2019. PMID: 31698727 Free PMC article. Review.
References
-
- Pestova T, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, editors. Translational Control in Biology and Medicine. Hershey JWB: Cold Sring Harbor Laboratory Press; 2007. pp. 87–128.
-
- Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, editors. Translational Control in Biology and Medicine. Hershey JWB: Cold Spring Harbor Laboratory Press; 2007. pp. 129–153.
-
- Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Hershey J: Cold Spring Harbor Laboratroy Press; 2007. pp. 155–172.
-
- Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231–1241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

