The fic domain: regulation of cell signaling by adenylylation
- PMID: 19362538
- PMCID: PMC2820730
- DOI: 10.1016/j.molcel.2009.03.008
The fic domain: regulation of cell signaling by adenylylation
Abstract
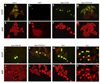
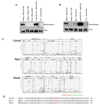
We show that the secreted antigen, IbpA, of the respiratory pathogen Histophilus somni induces cytotoxicity in mammalian cells via its Fic domains. Fic domains are defined by a core HPFxxGNGR motif and are conserved from bacteria to humans. We demonstrate that the Fic domains of IbpA catalyze a unique reversible adenylylation event that uses ATP to add an adenosine monophosphate (AMP) moiety to a conserved tyrosine residue in the switch I region of Rho GTPases. This modification requires the conserved histidine of the Fic core motif and renders Rho GTPases inactive. We further demonstrate that the only human protein containing a Fic domain, huntingtin yeast-interacting protein E (HYPE), also adenylylates Rho GTPases in vitro. Thus, we classify Fic domain-containing proteins as a class of enzymes that mediate bacterial pathogenesis as well as a previously unrecognized eukaryotic posttranslational modification that may regulate key signaling events.
Figures





References
-
- Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–13204. - PubMed
-
- Cole SP, Guiney DG, Corbeil LB. Two linked genes for outer membrane proteins are absent in four non-disease strains of Haemophilus somnus. Mol Microbiol. 1992;6:1895–1902. - PubMed
-
- Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. - PubMed
-
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

