Defining the structural basis of human plasminogen binding by streptococcal surface enolase
- PMID: 19363026
- PMCID: PMC2719351
- DOI: 10.1074/jbc.M109.004317
Defining the structural basis of human plasminogen binding by streptococcal surface enolase
Abstract
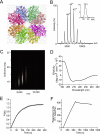
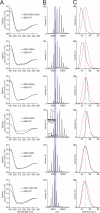
The flesh-eating bacterium group A Streptococcus (GAS) binds and activates human plasminogen, promoting invasive disease. Streptococcal surface enolase (SEN), a glycolytic pathway enzyme, is an identified plasminogen receptor of GAS. Here we used mass spectrometry (MS) to confirm that GAS SEN is octameric, thereby validating in silico modeling based on the crystal structure of Streptococcus pneumoniae alpha-enolase. Site-directed mutagenesis of surface-located lysine residues (SEN(K252 + 255A), SEN(K304A), SEN(K334A), SEN(K344E), SEN(K435L), and SEN(Delta434-435)) was used to examine their roles in maintaining structural integrity, enzymatic function, and plasminogen binding. Structural integrity of the GAS SEN octamer was retained for all mutants except SEN(K344E), as determined by circular dichroism spectroscopy and MS. However, ion mobility MS revealed distinct differences in the stability of several mutant octamers in comparison with wild type. Enzymatic analysis indicated that SEN(K344E) had lost alpha-enolase activity, which was also reduced in SEN(K334A) and SEN(Delta434-435). Surface plasmon resonance demonstrated that the capacity to bind human plasminogen was abolished in SEN(K252 + 255A), SEN(K435L), and SEN(Delta434-435). The lysine residues at positions 252, 255, 434, and 435 therefore play a concerted role in plasminogen acquisition. This study demonstrates the ability of combining in silico structural modeling with ion mobility-MS validation for undertaking functional studies on complex protein structures.
Figures


References
-
- Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. ( 2005) Lancet Infect. Dis. 5, 685– 694 - PubMed
-
- Tart A. H., Walker M. J., Musser J. M. ( 2007) Trends Microbiol. 15, 318– 325 - PubMed
-
- McArthur J. D., McKay F. C., Ramachandran V., Shyam P., Cork A. J., Sanderson-Smith M. L., Cole J. N., Ringdahl U., Sjöbring U., Ranson M., Walker M. J. ( 2008) FASEB J. 22, 3146– 3153 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

