Inter- and intraspecific variations of bacterial communities associated with marine sponges from san juan island, washington
- PMID: 19363076
- PMCID: PMC2687283
- DOI: 10.1128/AEM.00002-09
Inter- and intraspecific variations of bacterial communities associated with marine sponges from san juan island, washington
Abstract
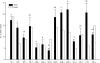
This study attempted to assess whether conspecific or congeneric sponges around San Juan Island, Washington, harbor specific bacterial communities. We used a combination of culture-independent DNA fingerprinting techniques (terminal restriction fragment length polymorphism and denaturing gradient gel electrophoresis [DGGE]) and culture-dependent approaches. The results indicated that the bacterial communities in the water column consisted of more diverse bacterial ribotypes than and were drastically different from those associated with the sponges. High levels of similarity in sponge-associated bacterial communities were found only in Myxilla incrustans and Haliclona rufescens, while the bacterial communities in Halichondria panicea varied substantially among sites. Certain terminal restriction fragments or DGGE bands were consistently obtained for different individuals of M. incrustans and H. rufescens collected from different sites, suggesting that there are stable or even specific associations of certain bacteria in these two sponges. However, no specific bacterial associations were found for H. panicea or for any one sponge genus. Sequencing of nine DGGE bands resulted in recovery of seven sequences that best matched the sequences of uncultured Proteobacteria. Three of these sequences fell into the sponge-specific sequence clusters previously suggested. An uncultured alphaproteobacterium and a culturable Bacillus sp. were found exclusively in all M. incrustans sponges, while an uncultured gammaproteobacterium was unique to H. rufescens. In contrast, the cultivation approach indicated that sponges contained a large proportion of Firmicutes, especially Bacillus, and revealed large variations in the culturable bacterial communities associated with congeneric and conspecific sponges. This study revealed sponge species-specific but not genus- or site-specific associations between sponges and bacterial communities and emphasized the importance of using a combination of techniques for studying microbial communities.
Figures




References
-
- Althoff, K., C. Schütt, A. Krasko, R. Steffen, R. Batel, and W. E. G. Müller. 1998. Evidence for a symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbor also for putatively toxic bacteria? Mar. Biol. 130:529-536.
-
- Arillo, A., G. Bacestrello, B. Burlando, and M. Sara. 1993. Metabolic integration between symbiotic cyanobacteria and sponges—a possible mechanism. Mar. Biol. 117:159-162.
-
- Beer, S., and M. Ilan. 1998. In situ measurements of photosynthetic irradiance responses of two Red Sea sponges growing under dim light conditions. Mar. Biol. 131:613-617.
-
- Bewley, C. A., N. D. Holland, and D. J. Faulkner. 1996. Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52:716-722. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

