Mitochondrial and nuclear localization of a novel pea thioredoxin: identification of its mitochondrial target proteins
- PMID: 19363090
- PMCID: PMC2689981
- DOI: 10.1104/pp.109.138073
Mitochondrial and nuclear localization of a novel pea thioredoxin: identification of its mitochondrial target proteins
Abstract
Plants contain several genes encoding thioredoxins (Trxs), small proteins involved in the regulation of the activity of many enzymes through dithiol-disulfide exchange. In addition to chloroplastic and cytoplasmic Trx systems, plant mitochondria contain a reduced nicotinamide adenine dinucleotide phosphate-dependent Trx reductase and a specific Trx o, and to date, there have been no reports of a gene encoding a plant nuclear Trx. We report here the presence in pea (Pisum sativum) mitochondria and nuclei of a Trx isoform (PsTrxo1) that seems to belong to the Trx o group, although it differs from this Trx type by its absence of introns in the genomic sequence. Western-blot analysis with isolated mitochondria and nuclei, immunogold labeling, and green fluorescent protein fusion constructs all indicated that PsTrxo1 is present in both cell compartments. Moreover, the identification by tandem mass spectrometry of the native mitochondrial Trx after gel filtration using the fast-protein liquid chromatography system of highly purified mitochondria and the in vitro uptake assay into isolated mitochondria also corroborated a mitochondrial location for this protein. The recombinant PsTrxo1 protein has been shown to be reduced more effectively by the Saccharomyces cerevisiae mitochondrial Trx reductase Trr2 than by the wheat (Triticum aestivum) cytoplasmic reduced nicotinamide adenine dinucleotide phosphate-dependent Trx reductase. PsTrxo1 was able to activate alternative oxidase, and it was shown to interact with a number of mitochondrial proteins, including peroxiredoxin and enzymes mainly involved in the photorespiratory process.
Figures

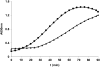
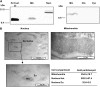

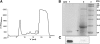
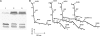
References
-
- Arai RJ, Masutani H, Yodoi J, Debbas V, Laurindo FR, Stern A, Monteiro HP (2006) Nitric oxide induces thioredoxin-1 nuclear translocation: possible association with the p21Ras survival pathway. Biochem Biophys Res Commun 348 1254–1260 - PubMed
-
- Arner ESJ, Holmgren A (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267 6102–6109 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

