Drosophila dosage compensation: a complex voyage to the X chromosome
- PMID: 19363150
- PMCID: PMC2674252
- DOI: 10.1242/dev.029645
Drosophila dosage compensation: a complex voyage to the X chromosome
Abstract
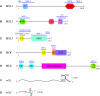
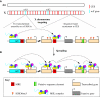
Dosage compensation is the crucial process that equalizes gene expression from the X chromosome between males (XY) and females (XX). In Drosophila, the male-specific lethal (MSL) ribonucleoprotein complex mediates dosage compensation by upregulating transcription from the single male X chromosome approximately twofold. A key challenge is to understand how the MSL complex distinguishes the X chromosome from autosomes. Recent studies suggest that this occurs through a multi-step targeting mechanism that involves DNA sequence elements and epigenetic marks associated with transcription. This review will discuss the relative contributions of sequence elements and transcriptional marks to the complete pattern of MSL complex binding.
Figures


References
-
- Akhtar, A. and Becker, P. B. (2000). Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367-375. - PubMed
-
- Akhtar, A., Zink, D. and Becker, P. B. (2000). Chromodomains are protein-RNA interaction modules. Nature 407, 405-409. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

