Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis
- PMID: 19363473
- PMCID: PMC2858974
- DOI: 10.1038/nature08012
Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis
Abstract
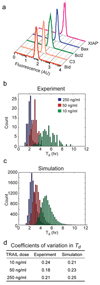
In microorganisms, noise in gene expression gives rise to cell-to-cell variability in protein concentrations. In mammalian cells, protein levels also vary and individual cells differ widely in their responsiveness to uniform physiological stimuli. In the case of apoptosis mediated by TRAIL (tumour necrosis factor (TNF)-related apoptosis-inducing ligand) it is common for some cells in a clonal population to die while others survive-a striking divergence in cell fate. Among cells that die, the time between TRAIL exposure and caspase activation is highly variable. Here we image sister cells expressing reporters of caspase activation and mitochondrial outer membrane permeabilization after exposure to TRAIL. We show that naturally occurring differences in the levels or states of proteins regulating receptor-mediated apoptosis are the primary causes of cell-to-cell variability in the timing and probability of death in human cell lines. Protein state is transmitted from mother to daughter, giving rise to transient heritability in fate, but protein synthesis promotes rapid divergence so that sister cells soon become no more similar to each other than pairs of cells chosen at random. Our results have implications for understanding 'fractional killing' of tumour cells after exposure to chemotherapy, and for variability in mammalian signal transduction in general.
Figures


Comment in
-
Of elections and cell-death decisions.Mol Cell. 2009 May 15;34(3):257-8. doi: 10.1016/j.molcel.2009.05.001. Mol Cell. 2009. PMID: 19450524 Free PMC article.
-
Systems biology: When it is time to die.Nature. 2009 May 21;459(7245):334-5. doi: 10.1038/459334a. Nature. 2009. PMID: 19458703 No abstract available.
References
-
- Blake WJ, AErn M K, Cantor CR, et al. Nature. 2003;422(6932):633. - PubMed
-
- Colman-Lerner A, Gordon A, Serra E, et al. Nature. 2005;437(7059):699. - PubMed
-
- Elowitz MB, Levine AJ, Siggia ED, et al. Science (New York, N.Y. 2002;297(5584):1183. - PubMed
-
- Golding I, Paulsson J, Zawilski SM, et al. Cell. 2005;123(6):1025. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

