Polyubiquitination by HECT E3s and the determinants of chain type specificity
- PMID: 19364824
- PMCID: PMC2698738
- DOI: 10.1128/MCB.00240-09
Polyubiquitination by HECT E3s and the determinants of chain type specificity
Abstract
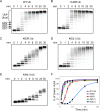
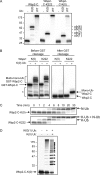
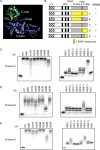
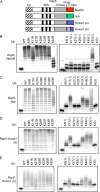
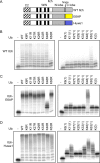
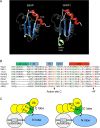
Polyubiquitination can mediate several different biochemical functions, determined in part by which lysine of ubiquitin is used to link the polyubiquitin chain. Among the HECT domain ubiquitin ligases, some, such as human E6AP, preferentially catalyze the formation of K48-linked polyubiquitin chains, while others, including Saccharomyces cerevisiae Rsp5 and human Itch, preferentially catalyze the formation of K63-linked chains. The features of HECT E3s that determine their chain type specificities have not been identified. We show here that chain type specificity is a function solely of the Rsp5 HECT domain, that the identity of the cooperating E2 protein does not influence the chain type specificity, that single chains produced by Rsp5 contain between 12 and 30 ubiquitin moieties, and that the determinants of chain type specificity are located within the last 60 amino acids of the C lobe of the HECT domain. Our results are also consistent with a simple sequential-addition mechanism for polyubiquitination by Rsp5, rather than a mechanism involving the formation of either E2- or E3-linked polyubiquitin chain transfers.
Figures



References
-
- Adhikary, S., F. Marinoni, A. Hock, E. Hulleman, N. Popov, R. Beier, S. Bernard, M. Quarto, M. Capra, S. Goettig, U. Kogel, M. Scheffner, K. Helin, and M. Eilers. 2005. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123409-421. - PubMed
-
- Bernassola, F., M. Karin, A. Ciechanover, and G. Melino. 2008. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 1410-21. - PubMed
-
- Bothos, J., M. K. Summers, M. Venere, D. M. Scolnick, and T. D. Halazonetis. 2003. The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene 227101-7107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
