Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine
- PMID: 19364832
- PMCID: PMC2708557
- DOI: 10.1128/IAI.00059-09
Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine
Abstract
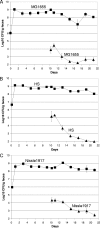
Different Escherichia coli strains generally have the same metabolic capacity for growth on sugars in vitro, but they appear to use different sugars in the streptomycin-treated mouse intestine (Fabich et al., Infect. Immun. 76:1143-1152, 2008). Here, mice were precolonized with any of three human commensal strains (E. coli MG1655, E. coli HS, or E. coli Nissle 1917) and 10 days later were fed 10(5) CFU of the same strains. While each precolonized strain nearly eliminated its isogenic strain, confirming that colonization resistance can be modeled in mice, each allowed growth of the other commensal strains to higher numbers, consistent with different commensal E. coli strains using different nutrients in the intestine. Mice were also precolonized with any of five commensal E. coli strains for 10 days and then were fed 10(5) CFU of E. coli EDL933, an O157:H7 pathogen. E. coli Nissle 1917 and E. coli EFC1 limited growth of E. coli EDL933 in the intestine (10(3) to 10(4) CFU/gram of feces), whereas E. coli MG1655, E. coli HS, and E. coli EFC2 allowed growth to higher numbers (10(6) to 10(7) CFU/gram of feces). Importantly, when E. coli EDL933 was fed to mice previously co-colonized with three E. coli strains (MG1655, HS, and Nissle 1917), it was eliminated from the intestine (<10 CFU/gram of feces). These results confirm that commensal E. coli strains can provide a barrier to infection and suggest that it may be possible to construct E. coli probiotic strains that prevent growth of pathogenic E. coli strains in the intestine.
Figures


References
-
- Anderson, J. D., W. A. Gillespie, and M. H. Richmond. 1973. Chemotherapy and antibiotic-resistance transfer between Enterobacteria in the human gastro-intestinal tract. J. Med. Microbiol. 6461-473. - PubMed
-
- Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, J. I. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, DC.
-
- Bergthorsson, U., and H. Ochman. 1998. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 156-16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

