Crystal structure of Bacillus anthracis dihydrofolate reductase with the dihydrophthalazine-based trimethoprim derivative RAB1 provides a structural explanation of potency and selectivity
- PMID: 19364848
- PMCID: PMC2704665
- DOI: 10.1128/AAC.01666-08
Crystal structure of Bacillus anthracis dihydrofolate reductase with the dihydrophthalazine-based trimethoprim derivative RAB1 provides a structural explanation of potency and selectivity
Abstract
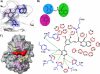
Bacillus anthracis possesses an innate resistance to the antibiotic trimethoprim due to poor binding to dihydrofolate reductase (DHFR); currently, there are no commercial antibacterials that target this enzyme in B. anthracis. We have previously reported a series of dihydrophthalazine-based trimethoprim derivatives that are inhibitors for this target. In the present work, we have synthesized one compound (RAB1) displaying favorable 50% inhibitory concentration (54 nM) and MIC (< or =12.8 microg/ml) values. RAB1 was cocrystallized with the B. anthracis DHFR in the space group P2(1)2(1)2(1), and X-ray diffraction data were collected to a 2.3-A resolution. Binding of RAB1 causes a conformational change of the side chain of Arg58 and Met37 to accommodate the dihydrophthalazine moiety. Unlike the natural substrate or trimethoprim, the dihydrophthalazine group provides a large hydrophobic anchor that embeds within the DHFR active site and accounts for its selective inhibitory activity against B. anthracis.
Figures

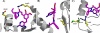
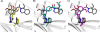
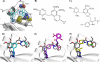
References
-
- Adams, P. D., K. Gopal, R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. K. Pai, R. J. Read, T. D. Romo, J. C. Sacchettini, N. K. Sauter, L. C. Storoni, and T. C. Terwilliger. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Sync. Rad. 11:53-55. - PubMed
-
- Adane, L., and P. V. Bharatam. 2008. Modelling and informatics in the analysis of P. falciparum DHFR enzyme inhibitors. Curr. Med. Chem. 15:1552-1569. - PubMed
-
- Appleman, J. R., W. A. Beard, T. J. Delcamp, N. J. Prendergast, J. H. Freisheim, and R. L. Blakley. 1990. Unusual transient- and steady-state kinetic behavior is predicted by the kinetic scheme operational for recombinant human dihydrofolate reductase. J. Biol. Chem. 265:2740-2748. - PubMed
-
- Baccanari, D. P., and L. F. Kuyper. 1993. Basis of selectivity of antibacterial diaminopyrimidines. J. Chemother. 5:393-399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information

