Architecture of complex I and its implications for electron transfer and proton pumping
- PMID: 19366614
- PMCID: PMC2699368
- DOI: 10.1016/j.bbabio.2009.01.012
Architecture of complex I and its implications for electron transfer and proton pumping
Abstract
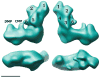
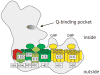
Proton pumping NADH:ubiquinone oxidoreductase (complex I) is the largest and remains by far the least understood enzyme complex of the respiratory chain. It consists of a peripheral arm harbouring all known redox active prosthetic groups and a membrane arm with a yet unknown number of proton translocation sites. The ubiquinone reduction site close to iron-sulfur cluster N2 at the interface of the 49-kDa and PSST subunits has been mapped by extensive site directed mutagenesis. Independent lines of evidence identified electron transfer events during reduction of ubiquinone to be associated with the potential drop that generates the full driving force for proton translocation with a 4H(+)/2e(-) stoichiometry. Electron microscopic analysis of immuno-labelled native enzyme and of a subcomplex lacking the electron input module indicated a distance of 35-60 A of cluster N2 to the membrane surface. Resolution of the membrane arm into subcomplexes showed that even the distal part harbours subunits that are prime candidates to participate in proton translocation because they are homologous to sodium/proton antiporters and contain conserved charged residues in predicted transmembrane helices. The mechanism of redox linked proton translocation by complex I is largely unknown but has to include steps where energy is transmitted over extremely long distances. In this review we compile the available structural information on complex I and discuss implications for complex I function.
Figures


References
-
- Brandt U. Energy converting NADH:quinone oxidoreductase (Complex I) Annu Rev Biochem. 2006;75:69–92. - PubMed
-
- Vinogradov AD. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (Complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim Biophys Acta. 1998;1364:169–185. - PubMed
-
- Smeitink J, Van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2:342–352. - PubMed
-
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. - PubMed
-
- Lenaz G, Bovina C, Castelluccio C, Fato R, Formiggini G, Genova ML, Marchetti M, Merlo Pich M, Pallotti F, Parenti Castelli G, Biagini G. Mitochondrial Complex I defects in aging. Mol Cell Biochem. 1997;174:329–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

