Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells
- PMID: 19366680
- PMCID: PMC2713521
- DOI: 10.1074/jbc.M901201200
Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells
Abstract
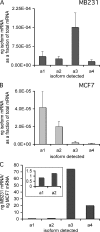
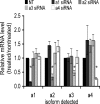
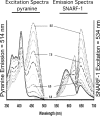
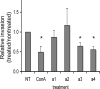
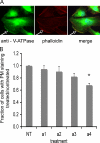
It has previously been shown that highly invasive MDA-MB231 human breast cancer cells express vacuolar proton-translocating ATPase (V-ATPases) at the cell surface, whereas the poorly invasive MCF7 cell line does not. Bafilomycin, a specific V-ATPase inhibitor, reduces the in vitro invasion of MB231 cells but not MCF7 cells. Targeting of V-ATPases to different cellular membranes is controlled by isoforms of subunit a. mRNA levels for a subunit isoforms were measured in MB231 and MCF7 cells using quantitative reverse transcription-PCR. The results show that although all four isoforms are detectable in both cell types, levels of a3 and a4 are much higher in MB231 than in MCF7 cells. Isoform-specific small interfering RNAs (siRNA) were employed to selectively reduce mRNA levels for each isoform in MB231 cells. V-ATPase function was assessed using the fluorescent indicators SNARF-1 and pyranine to monitor the pH of the cytosol and endosomal/lysosomal compartments, respectively. Cytosolic pH was decreased only on knockdown of a3, whereas endosome/lysosome pH was increased on knockdown of a1, a2, and a3. Treatment of cells with siRNA to a4 did not affect either cytosolic or endosome/lysosome pH. Measurement of invasion using an in vitro transwell assay revealed that siRNAs to both a3 and a4 significantly inhibited invasion of MB231 cells. Immunofluorescence staining of MB231 cells for V-ATPase distribution revealed extensive intracellular staining, with plasma membrane staining observed in approximately 18% of cells. Knockdown of a4 had the greatest effect on plasma membrane staining, leading to a 32% reduction. These results suggest that the a4 isoform may be responsible for targeting V-ATPases to the plasma membrane of MB231 cells and that cell surface V-ATPases play a significant role in invasion. However, other V-ATPases affecting the pH of the cytosol and intracellular compartments, particularly those containing a3, are also involved in invasion.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

