Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation
- PMID: 19366688
- PMCID: PMC2719359
- DOI: 10.1074/jbc.M808392200
Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation
Abstract
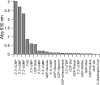
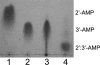
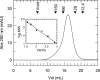
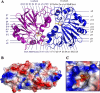
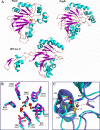
Carbon-phosphorus lyase is a multienzyme system encoded by the phn operon that enables bacteria to metabolize organophosphonates when the preferred nutrient, inorganic phosphate, is scarce. One of the enzymes encoded by this operon, PhnP, is predicted by sequence homology to be a metal-dependent hydrolase of the beta-lactamase superfamily. Screening with a wide array of hydrolytically sensitive substrates indicated that PhnP is an enzyme with phosphodiesterase activity, having the greatest specificity toward bis(p-nitrophenyl)phosphate and 2',3'-cyclic nucleotides. No activity was observed toward RNA. The metal ion dependence of PhnP with bis(p-nitrophenyl)phosphate as substrate revealed a distinct preference for Mn(2+) and Ni(2+) for catalysis, whereas Zn(2+) afforded poor activity. The three-dimensional structure of PhnP was solved by x-ray crystallography to 1.4 resolution. The overall fold of PhnP is very similar to that of the tRNase Z endonucleases but lacks the long exosite module used by these enzymes to bind their tRNA substrates. The active site of PhnP contains what are probably two Mn(2+) ions surrounded by an array of active site residues that are identical to those observed in the tRNase Z enzymes. A second, remote Zn(2+) binding site is also observed, composed of a set of cysteine and histidine residues that are strictly conserved in the PhnP family. This second metal ion site appears to stabilize a structural motif.
Figures


Similar articles
-
Structure and mechanism of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway.Biochemistry. 2011 Oct 11;50(40):8603-15. doi: 10.1021/bi2005398. Epub 2011 Sep 15. Biochemistry. 2011. PMID: 21830807
-
Physiological role of phnP-specified phosphoribosyl cyclic phosphodiesterase in catabolism of organophosphonic acids by the carbon-phosphorus lyase pathway.J Am Chem Soc. 2011 Mar 16;133(10):3617-24. doi: 10.1021/ja1102713. Epub 2011 Feb 22. J Am Chem Soc. 2011. PMID: 21341651
-
An unusual diphosphatase from the PhnP family cleaves reactive FAD photoproducts.Biochem J. 2018 Jan 11;475(1):261-272. doi: 10.1042/BCJ20170817. Biochem J. 2018. PMID: 29229761
-
Carbon-Phosphorus Lyase-the State of the Art.Appl Biochem Biotechnol. 2020 Apr;190(4):1525-1552. doi: 10.1007/s12010-019-03161-4. Epub 2019 Dec 2. Appl Biochem Biotechnol. 2020. PMID: 31792787 Review.
-
The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties.Biol Chem. 2005 Dec;386(12):1253-64. doi: 10.1515/BC.2005.142. Biol Chem. 2005. PMID: 16336119 Review.
Cited by
-
Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology.Curr Opin Chem Biol. 2013 Aug;17(4):580-8. doi: 10.1016/j.cbpa.2013.06.018. Epub 2013 Jul 17. Curr Opin Chem Biol. 2013. PMID: 23870698 Free PMC article. Review.
-
Metallo-β-lactamases in the Age of Multidrug Resistance: From Structure and Mechanism to Evolution, Dissemination, and Inhibitor Design.Chem Rev. 2021 Jul 14;121(13):7957-8094. doi: 10.1021/acs.chemrev.1c00138. Epub 2021 Jun 15. Chem Rev. 2021. PMID: 34129337 Free PMC article. Review.
-
Global regulation by the seven-component Pi signaling system.Curr Opin Microbiol. 2010 Apr;13(2):198-203. doi: 10.1016/j.mib.2010.01.014. Epub 2010 Feb 18. Curr Opin Microbiol. 2010. PMID: 20171928 Free PMC article. Review.
-
Microbial synthesis of pyrroloquinoline quinone.World J Microbiol Biotechnol. 2023 Dec 7;40(1):31. doi: 10.1007/s11274-023-03833-8. World J Microbiol Biotechnol. 2023. PMID: 38057682 Review.
-
Accumulation of intermediates of the carbon-phosphorus lyase pathway for phosphonate degradation in phn mutants of Escherichia coli.J Bacteriol. 2010 Jan;192(1):370-4. doi: 10.1128/JB.01131-09. J Bacteriol. 2010. PMID: 19854894 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

