Novel vascular endothelial growth factor D variants with increased biological activity
- PMID: 19366703
- PMCID: PMC2708897
- DOI: 10.1074/jbc.M109.001123
Novel vascular endothelial growth factor D variants with increased biological activity
Abstract
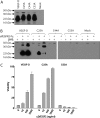
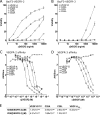
Members of the vascular endothelial growth factor (VEGF) family play a pivotal role in angiogenesis and lymphangiogenesis. They are potential therapeutics to induce blood vessel formation in myocardium and skeletal muscle, when normal blood flow is compromised. Most members of the VEGF/platelet derived growth factor protein superfamily exist as covalently bound antiparallel dimers. However, the mature form of VEGF-D (VEGF-D(DeltaNDeltaC)) is predominantly a non-covalent dimer even though the cysteine residues (Cys-44 and Cys-53) forming the intersubunit disulfide bridges in the other members of the VEGF family are also conserved in VEGF-D. Moreover, VEGF-D bears an additional cysteine residue (Cys-25) at the subunit interface. Guided by our model of VEGF-D(DeltaNDeltaC), the cysteines at the subunit interface were mutated to study the effect of these residues on the structural and functional properties of VEGF-D(DeltaNDeltaC). The conserved cysteines Cys-44 and Cys-53 were found to be essential for the function of VEGF-D(DeltaNDeltaC). More importantly, the substitution of the Cys-25 at the dimer interface by various amino acids improved the activity of the recombinant VEGF-D(DeltaNDeltaC) and increased the dimer to monomer ratio. Specifically, substitutions to hydrophobic amino acids Ile, Leu, and Val, equivalent to those found in other VEGFs, most favorably affected the activity of the recombinant VEGF-D(DeltaNDeltaC). The increased activity of these mutants was mainly due to stabilization of the protein. This study enables us to better understand the structural determinants controlling the biological activity of VEGF-D. The novel variants of VEGF-D(DeltaNDeltaC) described here are potential agents for therapeutic applications, where induction of vascular formation is required.
Figures






Similar articles
-
Adenoviral catheter-mediated intramyocardial gene transfer using the mature form of vascular endothelial growth factor-D induces transmural angiogenesis in porcine heart.Circulation. 2004 Mar 2;109(8):1029-35. doi: 10.1161/01.CIR.0000115519.03688.A2. Epub 2004 Feb 16. Circulation. 2004. PMID: 14967735
-
VEGF-D is the strongest angiogenic and lymphangiogenic effector among VEGFs delivered into skeletal muscle via adenoviruses.Circ Res. 2003 May 30;92(10):1098-106. doi: 10.1161/01.RES.0000073584.46059.E3. Epub 2003 Apr 24. Circ Res. 2003. PMID: 12714562
-
Angiogenic responses of vascular endothelial growth factors in periadventitial tissue.Hum Gene Ther. 2003 Oct 10;14(15):1451-62. doi: 10.1089/104303403769211664. Hum Gene Ther. 2003. PMID: 14577925
-
[Regulation of angiogenesis by VEGF signaling].Seikagaku. 2004 Dec;76(12):1534-42. Seikagaku. 2004. PMID: 15675367 Review. Japanese. No abstract available.
-
VEGF in physiological process and thyroid disease.Ann Endocrinol (Paris). 2007 Dec;68(6):438-48. doi: 10.1016/j.ando.2007.09.004. Epub 2007 Nov 7. Ann Endocrinol (Paris). 2007. PMID: 17991452 Review.
Cited by
-
Pulmonary Vasculopathy Associated with FIGF Gene Mutation.Am J Pathol. 2017 Jan;187(1):25-32. doi: 10.1016/j.ajpath.2016.09.008. Epub 2016 Nov 12. Am J Pathol. 2017. PMID: 27846380 Free PMC article.
-
Monomeric gremlin is a novel vascular endothelial growth factor receptor-2 antagonist.Oncotarget. 2016 Jun 7;7(23):35353-68. doi: 10.18632/oncotarget.9286. Oncotarget. 2016. PMID: 27174917 Free PMC article.
-
Multiplex detection of seven transgenes for human gene doping analysis.Sci Rep. 2025 Jun 20;15(1):20219. doi: 10.1038/s41598-025-06677-4. Sci Rep. 2025. PMID: 40542126 Free PMC article.
-
Clinical potential of angiogenic therapy and cellular reprogramming.JTCVS Open. 2021 Jun;6:108-115. doi: 10.1016/j.xjon.2020.12.023. Epub 2021 Mar 18. JTCVS Open. 2021. PMID: 34746874 Free PMC article.
-
Differential Receptor Binding and Regulatory Mechanisms for the Lymphangiogenic Growth Factors Vascular Endothelial Growth Factor (VEGF)-C and -D.J Biol Chem. 2016 Dec 30;291(53):27265-27278. doi: 10.1074/jbc.M116.736801. Epub 2016 Nov 16. J Biol Chem. 2016. PMID: 27852824 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

