Catalytic properties of RNase BN/RNase Z from Escherichia coli: RNase BN is both an exo- and endoribonuclease
- PMID: 19366704
- PMCID: PMC2708839
- DOI: 10.1074/jbc.M109.005462
Catalytic properties of RNase BN/RNase Z from Escherichia coli: RNase BN is both an exo- and endoribonuclease
Abstract
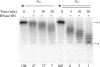
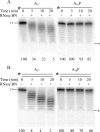
Processing of the 3' terminus of tRNA in many organisms is carried out by an endoribonuclease termed RNase Z or 3'-tRNase, which cleaves after the discriminator nucleotide to allow addition of the universal -CCA sequence. In some eubacteria, such as Escherichia coli, the -CCA sequence is encoded in all known tRNA genes. Nevertheless, an RNase Z homologue (RNase BN) is still present, even though its action is not needed for tRNA maturation. To help identify which RNA molecules might be potential substrates for RNase BN, we carried out a detailed examination of its specificity and catalytic potential using a variety of synthetic substrates. We show here that RNase BN is active on both double- and single-stranded RNA but that duplex RNA is preferred. The enzyme displays a profound base specificity, showing no activity on runs of C residues. RNase BN is strongly inhibited by the presence of a 3'-CCA sequence or a 3'-phosphoryl group. Digestion by RNase BN leads to 3-mers as the limit products, but the rate slows on molecules shorter than 10 nucleotides in length. Most interestingly, RNase BN acts as a distributive exoribonuclease on some substrates, releasing mononucleotides and a ladder of digestion products. However, RNase BN also cleaves endonucleolytically, releasing 3' fragments as short as 4 nucleotides. Although the presence of a 3'-phosphoryl group abolishes exoribonuclease action, it has no effect on the endoribonucleolytic cleavages. These data suggest that RNase BN may differ from other members of the RNase Z family, and they provide important information to be considered in identifying a physiological role for this enzyme.
Figures
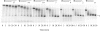
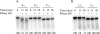



References
-
- Deutscher M. P. ( 1995) in tRNA: Structure, Biosynthesis, and Function ( Soll D., Rajbhandary U. L. eds) pp. 51– 65, American Society for Microbiology, Washington, D.C
-
- Deutscher M. P. ( 1990) Prog. Nucleic Acid Res. Mol. Biol. 39, 209– 240 - PubMed
-
- Vogel A., Schilling O., Späth B., Marchfelder A. ( 2005) Biol. Chem. 386, 1253– 1264 - PubMed
-
- Schierling K., Rösch S., Rupprecht R., Schiffer S., Marchfelder A. ( 2002) J. Mol. Biol. 316, 895– 902 - PubMed
-
- Redko Y., de la Sierra-Gallay I. L., Condon C. ( 2007) Nat. Rev. Microbiol. 5, 278– 286 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

